Figures & data
Figure 1. Correlation between NDT and RT+ for all ERV/LR families. Each data point represents a separate ERV/LR family. NDT is the normalized density of TFBS, and RT+ is the proportion of TFBS-containing elements in a family. “TFBS+/all” means RT+, and “TNT/all” means NDT.
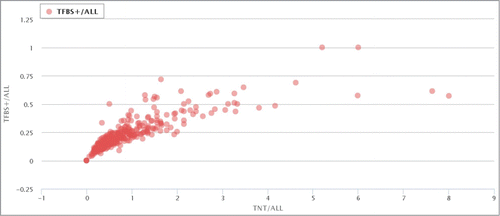
Figure 2. Distribution of the individual TFBS and DHS-containing ERV/LR elements is dependent on the number of TFBS per single element. “TFBS consisted HERV” means individual ERV/LR elements containing TFBS, shown in red; “TFBS & DNase I consisted HERV” means individual ERV/LR elements containing both TFBS and DNase I hypersensitivity site(s), shown in blue.
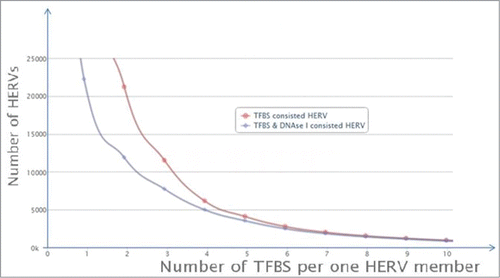
Table 1. List of the LTR5Hs elements selected for the experimental luciferase assay
Figure 3. Profiling of LTR enhancer activity in luciferase reporter experiments. (A) Schematic representation of the luciferase reporter constructs. Filled arrow – individual LTR elements tested in this assay; empty arrow – SV40 promoter; black bar – luciferase gene; (B) relative enhancer activities for LTR elements 1–12, established in a dual-luciferase assay. Data show means ± standard deviations of 4 independent experiments. Data is shown for the cell lines Tera-1, NT2/D1, A549, NGP127 and HepG2.
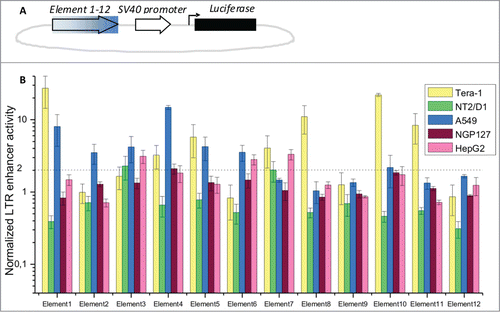
Figure 4. Proportion of TFBS-containing elements in correlation with the divergence of each ERV/LR family from their consensus sequences. Each data point represents a separate ERV/LR family. “TFBS+/all” means RT+. The divergence is shown as a millidiv score, with each unit equal to one substitution per 1000 nucleotides.
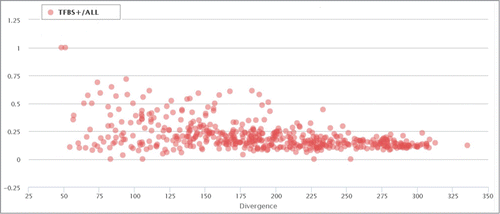
Figure 5. Distribution of TFBS for the particular transcription factor proteins among all the ERV/LR elements, in correlation with the divergence of the respective ERV/LR elements from their consensus sequence. The distribution is shown for NF-YA (red) and Rad21 (blue) transcription factor proteins. The divergence is shown as a millidiv score, with each unit equal to one substitution per 1000 nucleotides. The Y-axis is arbitrary and is customized for each transcription factor.
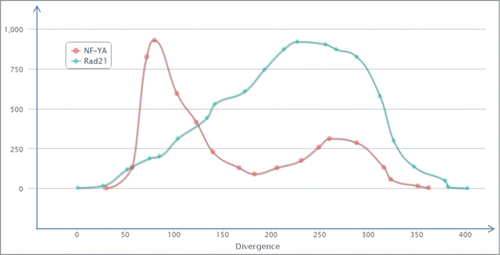
Figure 6. Screen shot of the representative HERV/LR browser output page. The user settings were “LTR5Hs” as the repeat family, “chr1” as the chromosome number. The Browser displays all the LTR5Hs inserts on the 1st chromosome, featuring TFBS – positive elements. An option is shown “show list of browser HERVs” that enables listing all the elements of a selected category on the browser screen, as a table supplemented by hyperlinks to the structure of particular each element. The zooming tool is enabled to facilitate navigation.
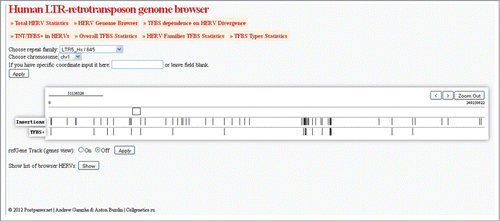
Table 2. Sequences of oligonucleotides used in this study
