Figures & data
Figure 1. Let-7a was downregulated in breast cancer and inversely correlated with KRas levels. (A) Representative examples of immunohistochemical staining for KRas in each of the clinical stages of breast cancers as indicated. (B) Quantification of KRas relative immunostaining intensity for each clinical stage of breast cancers. Data is shown as mean ± SEM for N as indicated in the figure in parenthesis as shown. (C) Relative expression levels of KRas mRNA in breast specimens. KRas abundance was normalized to 18S rRNA. Data is shown as mean ± SEM for N as indicated in the figure in parethesis as shown. (D) Relative expression levels of let-7a in human breast specimens. Let-7a(miRNA) levels were measured by TaqMan stem-loop qRT-PCR. U6 was used as the control. (E) Let-7a and KRas were inversely correlated in human breast cancers(r = −0.5355, p = 0.0023).
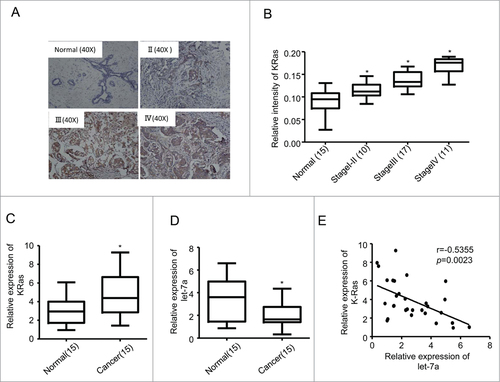
Figure 2. Let-7a inhibited mammosphere formation capacity in a KRas-dependent manner in breast cancer cells. (A) let-7a was detected after transfecting with lenti- let-7a and lenti-scramble, the fold-changes were showed as indicated(data are mean ± SD; n = 3). (B) Images showing mammosphere formation in MCF7 cells and SKBR3 cells transfected as indicated in (A). The cells were cultured with mammosphere culture medium containing EGF, bFGF and B27 supplement. Scale bar=100μm. (C and D) The efficiency of mammosphere-formation and the size of mammospheres were measured after 10 d in MCF7 cells and SKBR3 cells transfected lenti-let-7a and lenti-scramble or vector control (data are mean ± SD; n = 3, #P< 0.05, relative to the values in the respective controls). (E and F) MCF-7 and SKBR3 cells were infected with lenti-let-7 and/or KRas-siRNA. The number of mammospheres was counted, the percentage of mammosphere-forming cells was determined (data are mean ± SD; n = 3, #P < 0.05, relative to the values in the respective controls).
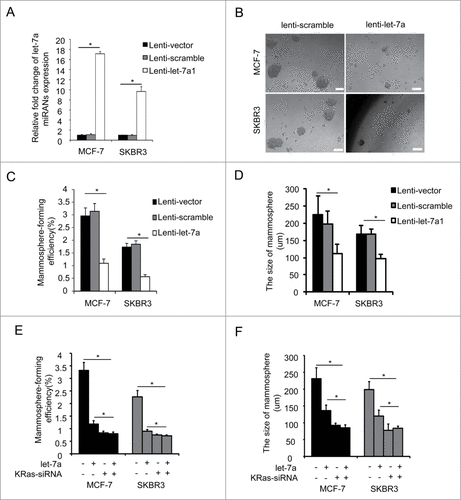
Figure 3. KRas is a direct target of let-7a. (A) The protein expression levels were determined in MCF-7 and SKBR3 cells transfected lenti-let-7a or lenti-scramble by western blot. (B-D) Luciferase activities were analyzed in 2 breast cell lines 36h after cotransfection of let-7a with either mutant or wild-type KRas luciferase reporter vectors.
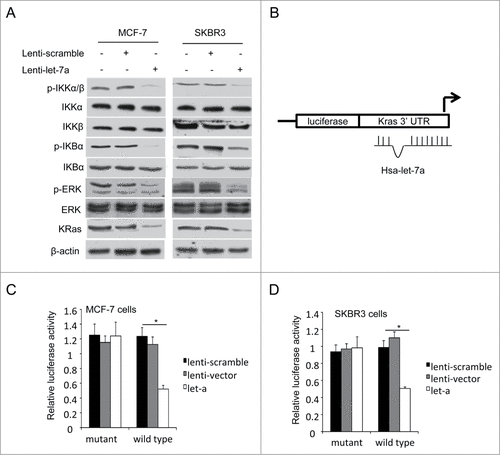
Figure 4. Let-7a regulates NF-κB and MAPK/ERK pathway in a KRas-dependent manner in breast cancer cells (A and B) The protein expression levels were determined in MCF-7 and SKBR3 cells transfected lenti- let-7a and/or KRas-siRNA by western blot. (C and D) The protein expression levels were determined in MCF-7 and SKBR3 cells transfected lenti- let-7a and/or KRas by western blot.
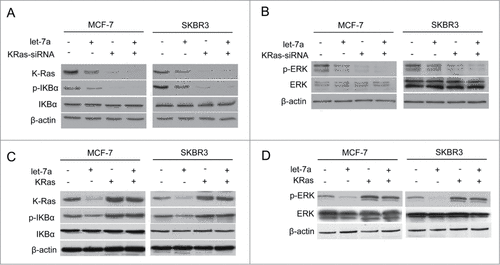
Figure 5 (See previous page). Let-7a/Ras/NF-κB axis inhibits mammosphere initiation. (A) Representative images of mammospheres in MCF-7 cell transfected or treated as indicated. Scale bar=100μm. (B and C) MCF-7 cells were infected with lenti-let-7 and/or KRas-siRNA, KRas or treated with 50μM SN50. The number of mammospheres was counted, the percentage of mammosphere-forming cells was determined (data are mean ± SD; n = 3, #P< 0.05, relative to the values in the respective controls). (D and E) The primary mammospheres of MCF-7 cells transfected lenti-let-7 and/or KRas-siRNA, KRas or treated with 50μM SN50 were dissociated into single cells and grown as secondary mammospheres. The size of mammospheres was measured after 10d (data are mean ± SD; n = 3, #P < 0.05, relative to the values in the respective controls). (F-I) FACS analysis was performed to identify the cells expressing ALDH activity. The proportion of ALDH+cells was measured(data are mean ± SD; n = 3, #P < 0.05, relative to the values in the respective controls). Representative images of the proportion of ALDH+cells in MCF-7 cell transfected or treated as indicated.
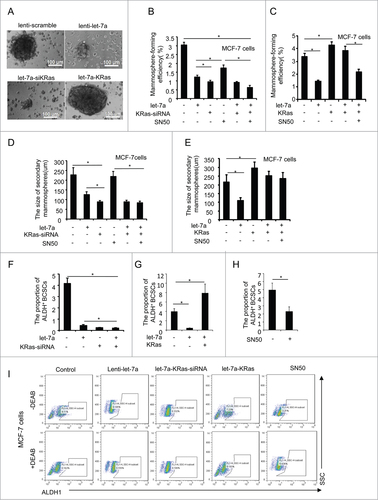
Figure 6. Let-7a affects the size of mammospheres through regulating Ras/MAPK/ERK pathway. (A and B) MCF-7 were infected with lenti-let-7 and/or KRas-siRNA, KRas or treated with 50 μM SN50. Relative cell viability was estimated at indicated time by a MTT assay in which each condition was replicated 3 times(data are mean ± SD, proliferation of untreated cells was defined as 100%). (C) MCF7 cells were treated with PD98059 (25μM). OD value was estimated at indicated time by a MTT assay in which was replicated 3 times(data are mean ± SD). (D and E) MCF-7 cells were transfected with lenti-let-7, and/or KRas-siRNA, KRas or treated with PD98059. The size of mammospheres was measured after 10d (data are mean ± SD; n = 3, #P< 0.05, relative to the values in the respective controls). (F) MCF-7 cells were treated with PD98059. The number of mammospheres was counted, the percentage of mammosphere-forming cells was determined (data are mean ± SD; n = 3, #P < 0.05, relative to the values in the respective controls). (G) Schematic overview of the effect of let-7a on mammosphere formation capacity.
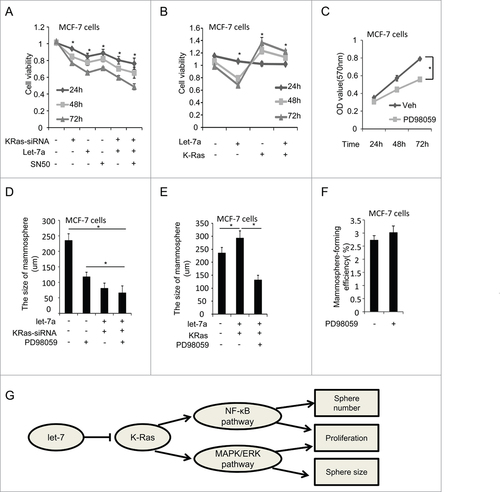
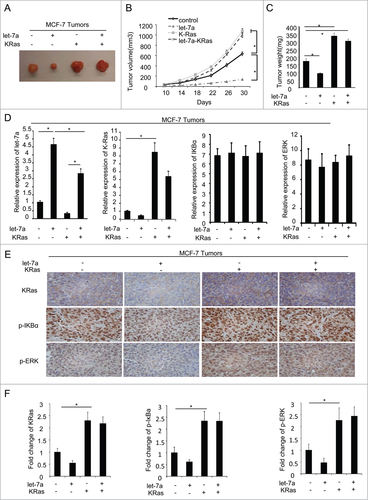
Figure 7. Let-7a inhibits tumor formation via KRas in vivo. MCF-7 cells transfected with lenti-let-7 and/or KRas(2×106 cells per site) mixed with matrigel (1:1) were injected into the mammary fat pad of immunodeficient mice supplemented with long-release estradiol pellets. Tumor size was measured every 4d until the tumors were harvested. The data was shown as mean ± SEM for N > 6 separate tumors for each group. (A) Images of tumors dissected from the mice. (B) The tumor size (mm3) versus days of post injection. (C) Tumor was weighted after resection at the end of experiment. (D) mRNA levels of let-7a, KRas, IκBα and ERK were determined by qRT-PCR in tumors. (E and F) IHC staining detected the protein level of KRas, p-IκBa and p-ERK in tumor tissues derived from mice. Data for quantified IHC was shown as mean ± SEM for N = 4 tumors in each group.