Figures & data
Figure 1. Effect of PGRN on glucose metabolism and insulin sensitivity, and its abrogation by PBA in vivo. All analyses compared age- and sex-matched mice fed a normal diet. Mice were distributed in 4 groups (12–15/per group): (1) vehicle (2) PGRN (i.p. 20 μg/day); (3) 4-PBA (orally, 1 g/Kg of body weight); (4) PGRN + 4-PBA. 12–15 mice per group were analyzed. (A) Effect of 3 weeks administration of rmPGRN and PBA on serum PGRN levels. (B) Body weight. (C) Blood glucose. (D) Serum insulin. (E) Serum triglyceride. (F) Energy expenditure. (G) Food intake. (H) GTT. (I) ITT. Data are means ± SD in each bar graph. *P < 0.05, **P < 0.01 (vs. vehicle). #P < 0.05 (vs. PGRN).
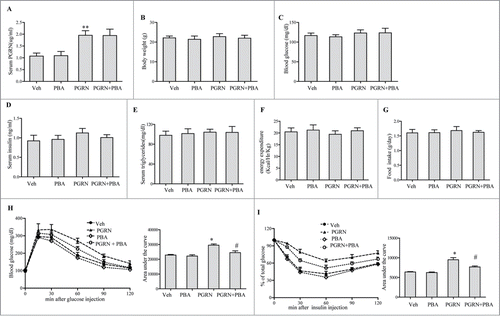
Figure 2. Effect of PGRN on lipid metabolism and liver steatosis in vivo. Mice were distributed as in . (A) β-oxidation of infused [1–14C] oleic acid in vivo. (B) Relative mRNA levels from genes associated with fatty acid and triacylglycerol (TG) synthesis, or β-oxidation in the liver. (C) Relative expression of genes related to β-oxidation in adipose tissue. (D) Oil Red O staining of liver sections (magnification 100×), scale bars: 20μm. (E) Mitochondria number and mitochondria area in liver. Data are means ± SEM in each bar graph. *P < 0.05, #P < 0.05 (vs. PGRN).
![Figure 2. Effect of PGRN on lipid metabolism and liver steatosis in vivo. Mice were distributed as in Figure 1. (A) β-oxidation of infused [1–14C] oleic acid in vivo. (B) Relative mRNA levels from genes associated with fatty acid and triacylglycerol (TG) synthesis, or β-oxidation in the liver. (C) Relative expression of genes related to β-oxidation in adipose tissue. (D) Oil Red O staining of liver sections (magnification 100×), scale bars: 20μm. (E) Mitochondria number and mitochondria area in liver. Data are means ± SEM in each bar graph. *P < 0.05, #P < 0.05 (vs. PGRN).](/cms/asset/cf1c0bc4-6714-4213-a625-1053431bb8d5/kccy_a_1041686_f0002_oc.gif)
Figure 3. Effects of PGRN on ER stress and insulin signaling in liver, adipose tissue and skeletal muscle. All analyses compared age- and sex-matched mice injected daily with saline solution or PGRN for 21 d. For insulin signaling, they were injected with 2 IU/kg of insulin (Ins). The relative quantity of proteins was analyzed using the Image J software. (A–C) Phosphorylation of IRS-1 tyrosine and Akt Ser-473 in liver, adipose tissue and skeletal muscle. (D–F) The phosphorylation of PERK and eIF2α in the liver, adipose tissue and skeletal muscle of mice co-treated with insulin and PGRN. The data expressed as means ± SEM in each bar graph. *P < 0.05, **P < 0.01 (vs. vehicle). #P < 0.05 (vs. Ins).
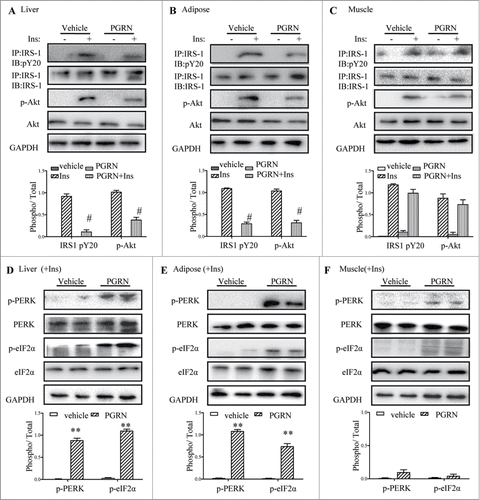
Figure 4. Effect of PBA treatment on markers of ER stress in the liver, adipose tissue, and muscle tissue. All analyses compare mice distributed in 4 groups (12–15/per group): (1) vehicle (2) PGRN (i.p. 20 μg/day); (3) 4-PBA (orally, 1 g/Kg of body weight); (4) PGRN + 4-PBA. For insulin signaling, they were injected with 2I U/Kg for insulin (Ins). (A–C) The indicators of ER stress and insulin receptor signaling were measured in liver, adipose and skeletal muscle at protein level. The data expressed as means ± SEM in each bar graph. *P < 0.05 (vs. vehicle). #P < 0.05 (vs. PGRN).
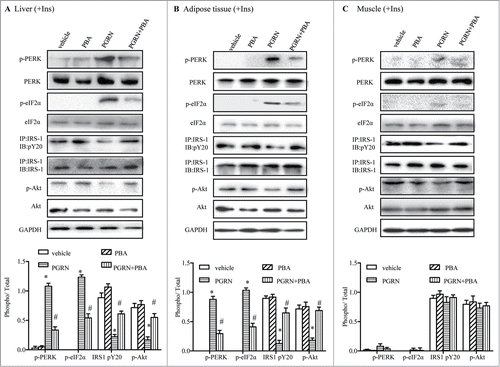
Figure 5. Time course of the trigger of ER stress and the inhibition of insulin signaling in vitro. Cells were treated with or without 100 ng/ml PGRN at indicated times. The indicators of ER stress and insulin receptor signaling were measured at protein level. The relative quantity of proteins was analyzed using the Image J software. (A–C) The phosphorylation of IRS-1 and Akt in hepatocytes, adipocytes and L6 cells. (D–F) The phosphorylation of PERK and eIF2α in hepatocytes, adipocytes and L6 cells. A representative blot is shown and data expressed as means ± SEM in each bar graph represent the average of 3 independent experiments. *P < 0.05, #P < 0.01 (vs. cells at time point of 0 minutes).
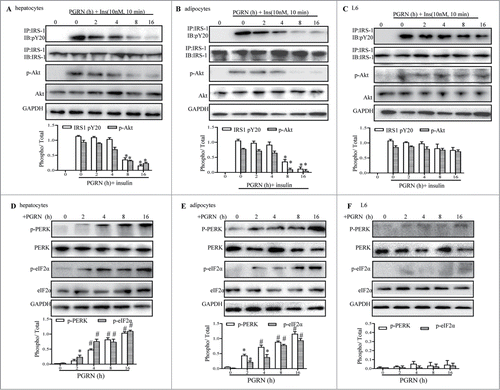
Figure 6. Involvements of PGRN in palmitate (palm)-induced IR in vitro. Control cells and PGRN-/- cells were pre-incubated with 0.5 mM palmitate for 16 h prior to incubation with 10 nM insulin (Ins) for 10 min. (A–C) PGRN content in culture media of cells with or without palmitate treatment. (D) The phosphorylation of IRS-1, Akt, PERK, and eIF2α in hepatocytes. (E) The phosphorylation of IRS-1, Akt, PERK, and eIF2α in adipocytes. (F) The phosphorylation of IRS-1, Akt, PERK, and eIF2α in L6 cells. The right is the quantification of proteins phosphorylation with normalization to total protein levels for each molecule under this condition. A representative blot is shown and all graphs show means ± SEM from at least 3 independent experiments. *P < 0.05, **P < 0.01 vs. the control groups. #P < 0.05 vs. the palmitate-treated groups.
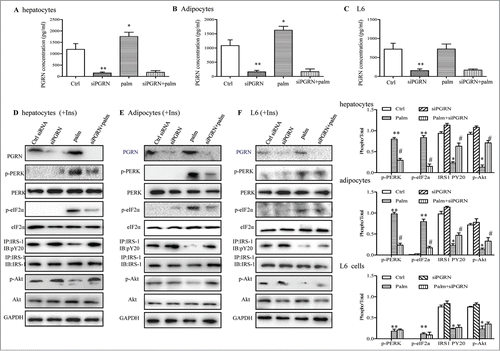
Figure 7. Knockdown of the PERK-eIF2α pathway blocks insulin resistance induced by PGRN in vitro. (A) Expression of PERK in hepatocytes. (B) Expression of PERK in adipocytes. Posphorylated eIF2α, total eIF2α, phosphorylated IRS-1 and total IRS-1 of PERK knockdown hepatocytes (C) and adipocytes (D) treated with PGRN (100 ng/ml) for 16 hr. The bottom is the quantification of proteins phosphorylation with normalization to total protein levels for each molecule under this condition. A representative blot is shown and all the data expressed as means ± SEM in each bar graph represent the average of at least 3 independent experiments. *P < 0.05 vs. the control siRNA groups. #P < 0.05 vs. the control siRNA + PGRN groups.
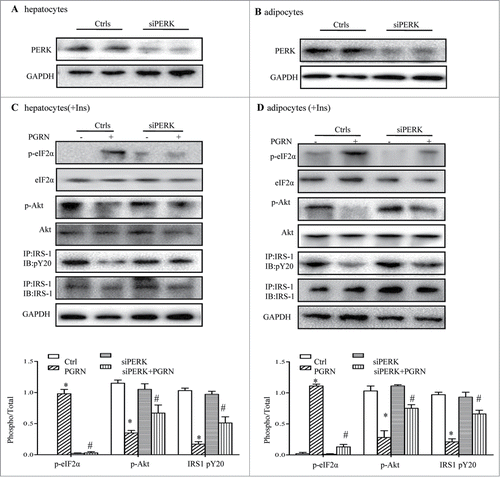
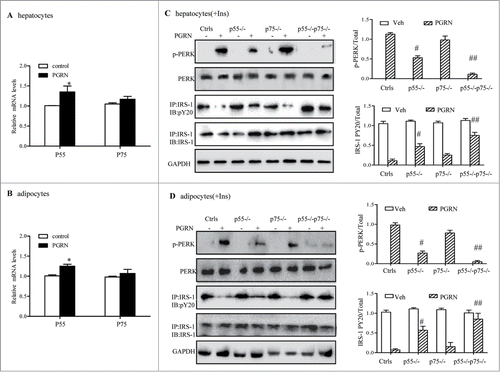
Figure 8. Improvement of PGRN-induced ER stress and insulin-stimulated IRS-1 phosphorylattion by blockade of TNFR in vitro. WT and TNFR1/2−/- cells were treated with vehicle or PGRN (100 ng/ml) for 16 hr. For insulin signaling, cells were stimulated with 10 nM of insulin (Ins) for 10 minutes. Indicators of ER stress and insulin signaling were measured at protein levels. The relative quantity of proteins was analyzed with Quantity One software. (A, B) Relative expression of TNFR1 (p55) receptor and TNFR2 (p75) in hepatocytes and adipocytes treated with PGRN (100 ng/ml) for 16 hr normalized to β-actin (real-time PCR). (C, D) phosphorylated PERK, total PERK, phosphorylated IRS-1 and total IRS-1 of WT, p55−/−, p75−/−, and p55−/− p75−/− hepatocytes and adipocytes. The right is the quantification of proteins phosphorylation with normalization to total protein levels for each molecule under this condition. The data expressed as means ± SEM in each bar graph represent the average of 3 independent experiments. *P < 0.05 vs. vehicle. #P < 0.05, #P < 0.01 vs. PGRN. IB, immunoblotting; IP, immunoprecipitation.