Figures & data
Figure 1. Microarray, qRT-PCR validation and the pipeline of the experimental strategy. (A) HepG2 cells were transfected with the scramble or CRT siRNA (10 nM) for 24 h. On termination of incubation, CRT levels were estimated by qRT-PCR using CRT specific primers. (B) RNA as in “A” was subjected to microarray analyses using Illumina Human HT-12 V3 expression Bead chip arrays. Genes differentially regulated (p < 0.5) are represented as black triangles in a volcano plot with the up- and downregulated genes being represented on either side of the central axis. (C) RNA from scramble and CRT siRNA treated HepG2 cells were subjected to qRT-PCR toward validation of differentially altered genes as obtained from the microarray. All fold changes were significant at p < 0.05. (D) Computational and experimental pipeline of the strategy followed for the altered genes. Values presented are means ±SEM of 3 experiments. ***p < 0.001 as compared to scramble transfected cells.
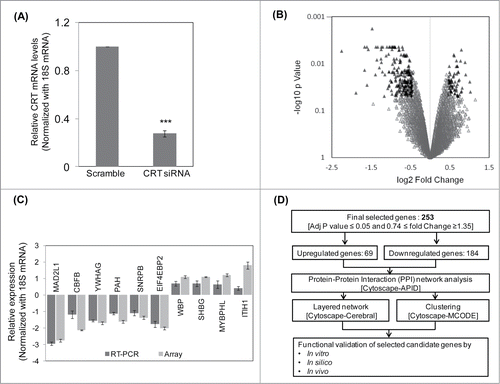
Figure 2. Protein-protein interaction (PPI) networks and sub-networks formed by the altered genes. (A) The set of 253 unique differentially expressed genes were assessed for their interacting partners using the APID2NET plugin in Cytoscape. The resultant PPI network composed of 818 nodes and 2794 edges and was used to generate a layered network based on subcellular localization of the 818 proteins. Blue colored nodes are representative of proteins encoded by the differentially expressed genes while the gray ones are the interacting partners. (B) The PPI as in ‘A’ was used to identify sub-networks using the MCODE clustering algorithm and the top 3 node-dense networks are shown.
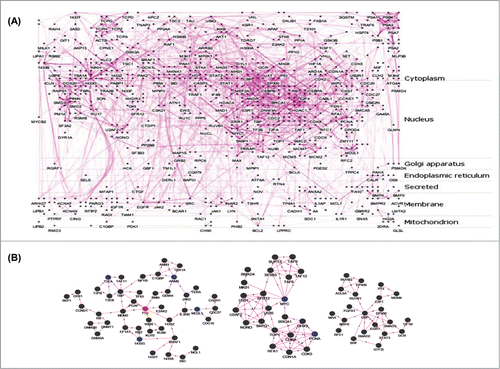
Figure 3. Calreticulin (CRT) inhibition down-regulates p53 and promotes lipid accumulation through up-regulation of lipogenic factors in HepG2 cells. (A) HepG2 cells were transfected with the scramble or CRTsiRNA (10 nM) for 48 h and on termination of incubation, cells were collected, lysed and lysates (30 µg) were assessed for the levels of p53 by western blot analysis. β-actin was taken as the loading control. (B) HepG2 cells were transfected with the scramble or p53 siRNA (10 nM) for 24 h. On termination of incubation, p53 levels were evaluated by qRT-PCR and normalized to the levels of 18S rRNA. (C) HepG2 cells transfected with either the scramble or p53 siRNA or CRT siRNA (10 nM) for 48 h. Cells were then fixed and stained with DAPI and Bodipy and viewed under a fluorescent microscope (63X). Bodipy fluorescence was quantified using the ImageJ software and these are given in the right panel of image (a.u). All experiments were done in triplicate and values in the histograms are means ±SEM. (D) Scramble, CRT siRNA and p53 siRNA transfected cells were assessed for the levels of the lipogenic genes: FAS and SREBP-1c by Western Blot analyses. Vinculin was taken as the loading control. Lower panel represents the densitometric analyses of respective blots normalized with loading control. Values are means ± SEM of 3 independent experiments. ***p < 0.001, **p < 0.01 and *p<0.05 as compared to scramble transfected cells.
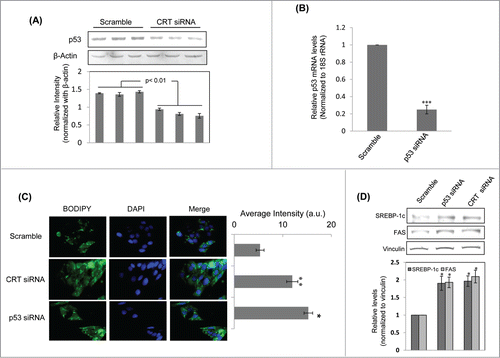
Figure 4. p53 binding site on the SREBP-1c promoter and its occupancy regulates SREBP-1c promoter activity. (A) The wild type (WT) SREBP-1c promoter harbouring 2 binding sites for p53 (site 1 at −219 bp and site 2 at −517bp) is shown. Mutations in both these sites were done independently by base substitution at 4 positions within the binding site (site 1 mutation: Mut1; site 2 mutation: Mut 2). (B) HepG2 cells were transfected with either the wild type SREBP-1c promoter construct (WT) or the mutated constructs (Mut1 or Mut2) and luciferase activity was assessed using the Dual luciferase assay kit. Renilla luciferase (30 ng) was cotransfected and used as the normalization control. (C) HepG2 cells were transfected with the WT or mutated SREBP-1c promoter constructs as in ‘B’ and also co-transfected with either the scramble or p53 siRNA or CRT siRNA or with CRT siRNA and the p53 cDNA clone (1 μg). After 48 h, cells were lysed and luciferase activities were determined. All experiments were done thrice and values are means ± SEM. **p < 0.01 and *p < 0.05 as compared to WT or Mut2 and #p < 0.05 as compared to CRT siRNA alone.
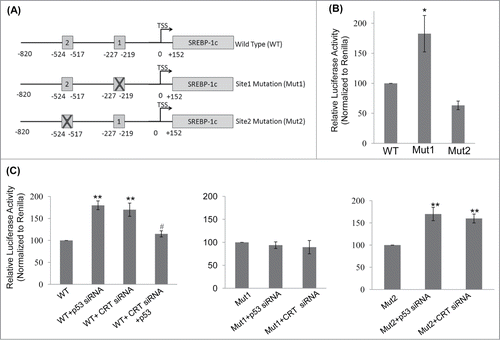
Figure 5. Binding of p53 to the SREBP-1c promoter and its decrease by CRT inhibition. (A) Double stranded oligonucleotides spanning the p53 binding site on the SREBP-1c promoter (site1) were labeled with γ-32P and incubated with purified p53 at 37°C for 30 min. On termination of incubation, samples were separated on a 5% polyacrylamide gel, dried and autoradiographed. A cold chase with unlabelled oligonucleotides at dilutions of 1:1 and 1:10 was done to check the specificity of the binding. (B) HepG2 cells were transfected with scramble or CRT siRNA (10 nM) for 48 h and the chromatin was isolated, sheared, immunoprecipitated with IgG or anti-p53 antibody and pulled down using protein A-sepharose beads. Immunoprecipitated DNA was quantified (qRT-PCR) for p53 enrichment on site1 using site specific primers as given in . Data was normalized to the IgG pulldown DNA. Experiments were done thrice and values are mean ±SEM. (C) HepG2 cells were transfected with CRT siRNA and co-transfected with p53 cDNA clone and incubated for 48h. On termination of incubations, cells were lysed and the levels of SREBP-1c were evaluated by protein gel blot analysis. β-Actin was taken as the loading control. Densitometric analyses are given in the panel below. (D) HepG2 cells were incubated in the absence or presence of palmitate or oleate for 24h and total RNA was isolated. CRT transcript levels were evaluated by qRT-PCR and data was normalized to 18S rRNA. All values are means±SEM and derived from 3 independent experiments. *p < 0.05 as compared to free probe (A); ***p < 0.001 as compared to scramble (B); ###p < 0.001 and #p < 0.05 as compared to control cells; ap < 0.05 as compared to CRT siRNA transfected cells.
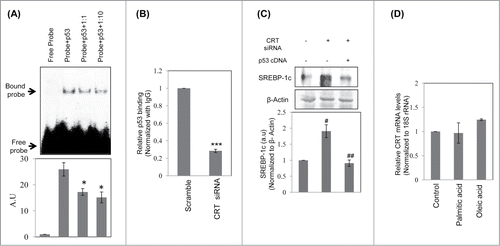
Table 1. List of primers
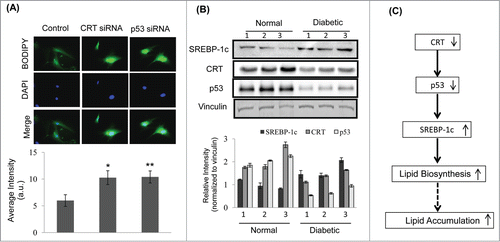
Figure 6. CRT downregulation promotes lipid accumulation in mice primary hepatocytes and its levels are inhibited in the db/db mice liver (A) Mice primary hepatocytes were transfected with either the scramble or CRT siRNA or p53 siRNA and stained with Bodipy and DAPI. Bodipy fluorescence was quantified and the data are given below. (B) 30 µg protein from normal and db/db mice (n = 3 ) liver were run on SDS-PAGE, transferred to nitrocellulose membranes and probed with antibodies against SREBP-1c, CRT and p53. Vinculin was used as the loading control and densitometric analyses of the same are given below. Experiment from each animal was run thrice and a representative blot is shown. (C) Schema demonstrating that CRT inhibition leads to p53 downregulation, that due to its decreased occupancy of the SREBP-1c promoter, facilitates its upregulation and subsequently promotes lipid synthesis and accumulation. All values are means±SEM from 3 independent experiments. **p < 0.01 and *p < 0.05 as compared to scramble transfected cells.