Figures & data
Figure 1. Full-length hCAP-H2 and its variant detected in human cell lines. (A) hCAP-H2 proteins in RPE-1, HeLa (cervical cancer), HCT116 (colon cancer), U2OS (osteosarcoma), HOSE4, and ovarian cancer cell lines (PEO-1, OVCAR5, OVCAR10, and SKOV3) were assessed by immunoblot analysis. Arrows indicate the full-length hCAP-H2 and its smaller variant. Tubulin serves as a loading control. (B) Asynchronous (Asy) and mitotic (M) RPE-1 and HeLa cells were subjected to Western blot analysis to detect hCAP-H2 proteins. Mitotic cells were prepared by mitotic shake-off. Cell lysates were treated with lambda protein phosphatase (λPPase). (C) Schematic protocol for the inhibitor treatments employed in panel D. (D) RPE-1 cells grown in normal medium were subsequently cultured for 48 hours in starvation medium, which consists of 20 times less fetal bovine serum (FBS) than normal. CDK4/6 inhibitors (Palbociclib and LEE011), DNA replication inhibitors (HU and Thymidine), or DMSO (control) were added to starvation medium 2 hours before serum re-stimulation. Cells were further cultured for 24 hours in normal medium (serum +) containing the inhibitors, and lysates were subjected to immunoblot analysis. P indicates phosphorylated Rb proteins. (E) The indicated cell lines were cultured for 2 days in normal (starvation −) or starvation medium (starvation +), followed by Western blot analysis. (F) RPE-1 cells grown in normal medium (C, control) were subsequently cultured for 2 days in starvation medium (S, Starvation). After the starvation treatment, cells were further cultured in normal medium for the indicated periods of time (Re-stimulation). Cell lysates derived from the respective culturing steps were subjected to immunoblot analysis. Band intensities (top) and expression ratio (bottom) of the hCAP-H2 isoforms are shown.
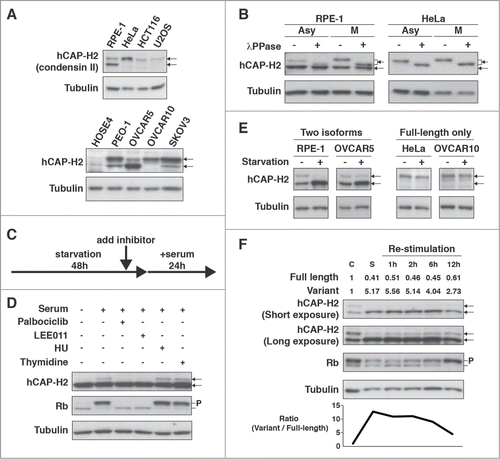
Figure 2. Role of the uORF in expression of the hCAP-H2ΔN variant. (A) The nucleotide sequence of the 5′ region of NCAPH2 mRNA. The small upstream ORF (uORF) contains start codon (uAUG, red) and stop codon (UGA, blue). The first AUG (AUG1) and second AUG (AUG2), responsible for translational initiation of hCAP-H2, are also highlighted in red. (B) Prediction of translational start sites for hCAP-H2. The NetStart 1.0 server was used for a prediction of translation start. Probability of translational initiation is scored between 0 and 1, and more than 0.5 represents typical start sites. (C) Schematic representation of the NCAPH2 constructs (wild type, ΔuAUG, ΔAUG1, and ΔAUG2). Expression of hCAP-H2 proteins fused to EYFP is under control of the CMV promoter. The mutated start codons are shown in red. (D) The NCAPH2 constructs shown in panel C were transfected into RPE-1 cells. Negative control (N.C.) indicates no transfection. hCAP-H2 proteins fused to EYFP were detected by Western blotting using hCAP-H2 antibody. An asterisk indicates an additional band observed in cells carrying the ΔAUG2 construct. (E) The full-length NCAPH2 (FL) and ΔN genes starting from the AUG1 and AUG2 codons, respectively, were cloned into pCMV6-Entry plasmids and expressed in RPE-1 cells. N.C. indicates RPE-1 cells without exogenous expression of the NCAPH2 genes. Cell lysates from RPE-1 and HeLa cells were subjected to immunoblot analysis. (F) Co-IP analysis testing the interaction between hCAP-H2 (FL, ΔN, and ΔC) and the other components of the condensin complex. hCAP-H2ΔC lacks the 130 a.a. of the hCAP-H2 C-terminus. Expression of Flag-tagged hCAP-H2 proteins was induced by doxycycline (Materials and Methods).
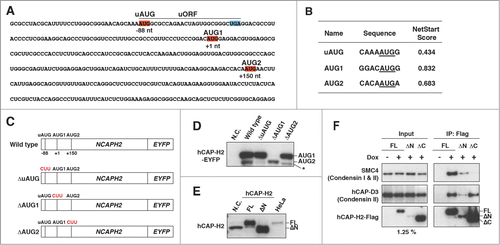
Figure 3. Overexpression of hCAP-H2 induces formation of DAPI-dense chromatin foci. (A) hCAP-H and hCAP-H2 (FL and ΔN) proteins fused to the Flag epitope were expressed in RPE-1 cells for 2 days and visualized by IF analysis. Scale bar indicates 10 µm. (B) Histone H3 di- and tri-methylated Lys9 were visualized by IF in RPE-1 cells expressing the full-length hCAP-H2. Inset shows enlarged views. Scale bar indicates 5 µm. (C) hCAP-H2 (FL and ΔN) proteins fused to the Flag epitope were expressed in RPE-1 cells and visualized by IF. These microscopic images, at a relatively high resolution, were captured by a Leica SP5 II laser scanning confocal microscope. Scale bar indicates 1 µm. (D) Chromatin-unbound (Fr. 1), DNase-extractable (Fr. 2), high salt-extractable (Fr. 3), and high salt-resistant (Fr. 4) fractions were prepared from asynchronous (Asy) and mitotic (M) RPE-1 cells (See Materials and Methods) and subjected to Western blotting. WCL indicates whole cell lysate. (E and F) The same fractionation was performed using RPE-1 cells expressing Flag-tagged hCAP-H2 proteins (FL and ΔN). The fractions were subjected to immuno blotting (E). The high salt-resistant fraction was further applied for IF analysis to co-visualize hCAP-H2 and lamin A/C proteins (F). Scale bar indicates 5 µm. (G) RPE-1 cells were transfected with plasmids encoding EYFP-tagged hCAP-H2 proteins (FL and ΔN) and culture for 2 days. Cells were subjected to IF analysis to stain CENP-B proteins, which bind to centromeric satellites. RPE-1 cells were infected with a retrovirus encoding H-RasV12 (Ras) or a control virus without H-RasV12 (Con) and subjected to IF analysis 8 days after the infection. Scale bar indicates 5 µm.
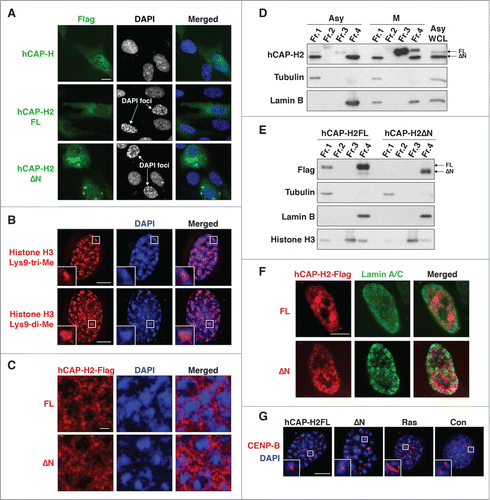
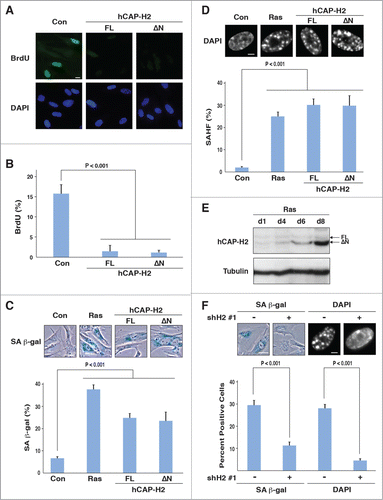
Figure 4. hCAP-H2 facilitates cellular senescence. (A and B) IMR90 cells were infected with retroviruses encoding the full-length hCAP-H2 or ΔN variant, or a control virus without encoding these proteins. BrdU incorporation and visualization were performed 8 days after the infection (A). Scale bar indicates 10 µm. Percentages of IMR90 cells showing BrdU staining were scored 8 days after retrovirus infection (B). (C and D) IMR90 cells were infected by retroviruses encoding the H-RasV12, full-length hCAP-H2, or ΔN variant, or a control virus, and subjected to SA-β-gal staining 8 days after the infection (C). Infected cells were stained by DAPI to assess SAHF formation (D). Scale bars in panels C and D indicate 50 µm and 5 µm, respectively. (E) hCAP-H2 levels were monitored from day 1 (d1) to day 8 (d8), after IMR90 cells were infected with a retrovirus encoding H-RasV12. Tubulin serves as a loading control. (F) IMR90 cells were infected with a retrovirus encoding H-RasV12 as well as a lentivirus encoding shRNA against NCAPH2 mRNA (shH2#1), and assessed for SA-β-gal activity and SAHF formation 8 days after the infection. Scale bars indicate 50 (left) and 5 µm (right).