Figures & data
Figure 1. Effect of cAMP on growth and cell size in cyr1Δ msn2Δ msn4Δ pde2Δ mutant. Asynchronous culture of the GG104 strain growing exponentially (1.7–2 × 106 cells/ml) in YPD medium was splitted into 2 fractions and 2mM cAMP was added to one of them (▪), while the other was the untreated control (♦). The black arrows indicate the time of cAMP addition. Samples were harvested every 30 minutes to measure the cell number (panel A), the absorbance at 600 nm (panel B), the percentage of budded cells (panel C) and the mean cell size (panel D).
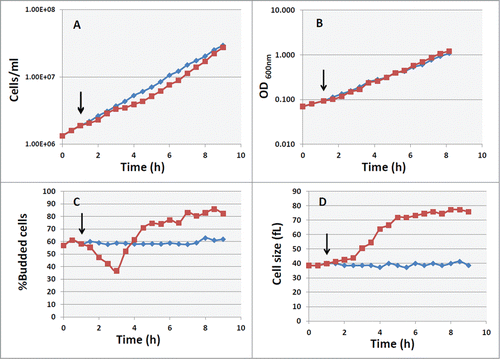
Figure 2. Protein and DNA distribution of GG104 cells. Histograms of protein and DNA distributions of control cultures and of cultures treated with 2 mM cAMP collected at the indicated time after cAMP addition. Yeast cells were collected fixed with 70% ethanol, stored at 4°C, and subsequently processed for flow cytometry, using a Becton Dickinson FACStarPlus. Fluorescein isothiocianate (FITC) for protein staining and Propidium Iodide (PI) for DNA staining were used. Approx 30.000 events were analyzed for each sample. Plot generation and analysis were performed with WinMDI2.9 software.
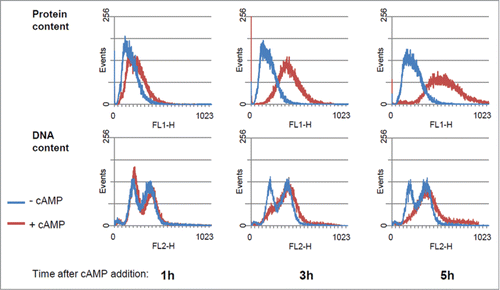
Table 1 Growth and Cell cycle parameters for GG104 strain growing without cAMP and 5 hours after addition of cAMP.
Figure 3. Evaluation of cells size at budding (Vs) and at cell division (Vm). GG104 cells exponentially growing were treated with 2 mM cAMP, stained with DAPI and photographed with a fluorescence microscope. Panel A: sample image of the cells growing without cAMP and after 3.5 hr of cAMP treatment. The image is taken in light transmission merged with DAPI fluorescence. Panel B: Cell size at budding (Vs) and at cell division (Vm) measured on the photographs taken at different times after cAMP addition. The average size of at least 30 cells with small buds (Vs) and of at least 30 binucleate cells (Vm) is reported. Error bars indicate Standard Deviation. *p < 0.05, **p < 0.01 is related to a comparison with the preceding mean value.
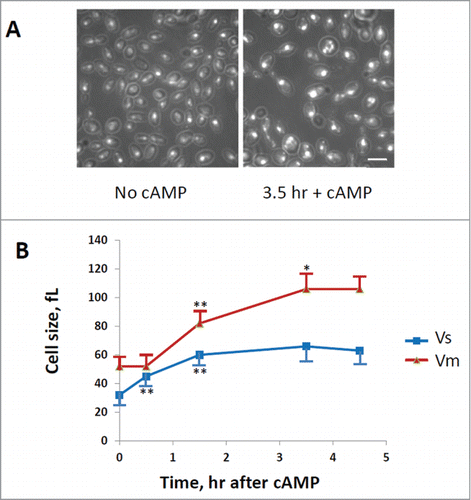
Figure 4. CLN1 but not CLN2 is required for G1 delay induced by cAMP addition. Asynchronous cultures of the GG104 cln1Δ (A) and GG104 cln2Δ (B) strains growing exponentially (1.7–2 × 106 cells/ml) in YPD medium were splitted into 2 fractions and 2mM cAMP was added to one of them (▪), while the other was the untreated control (♦). Samples were harvested every 30 minutes to assay cell number, cell size and budding index.
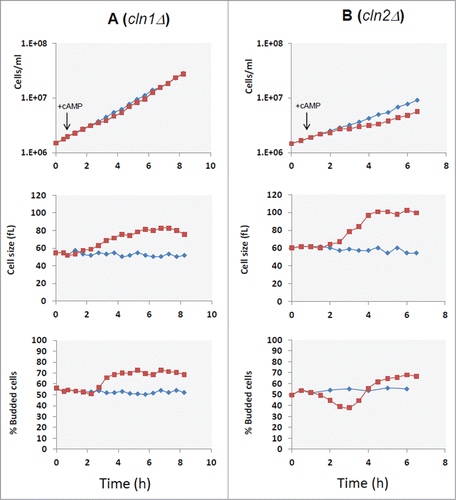
Figure 5. Cell size at budding and at cell division for cln1Δ strain. GG104 cln1Δ cells exponentially growing were treated with 2 mM cAMP, stained with DAPI and photographed with a fluorescence microscope. Cell size at budding (Vs) and at cell division (Vm) were measured on the images taken at different times after cAMP addition. The average size of at least 30 cells with small buds (Vs) and of at least 30 binucleate cells (Vm) is reported. Error bars indicate Standard Deviation. * p < 0.05, ** p < 0.01 is related to a comparison with the preceding mean value.
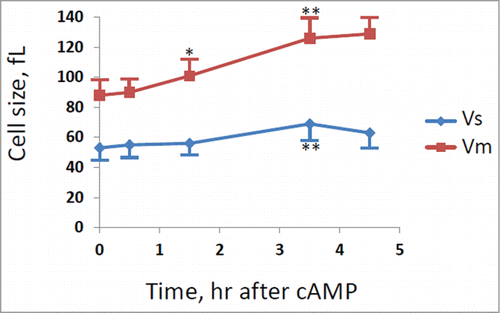
Figure 6. cAMP addition caused a CLN1 repression. Quantitative RealTime-PCR assays on CLN1 (hatched bars) and CLN2 (white bars) mRNA prepared from samples obtained during kinetic growth analysis, collected at the indicated time after cAMP addition. Graphic represents the mean values of 3 independent experiments, error bars indicate standard deviations.
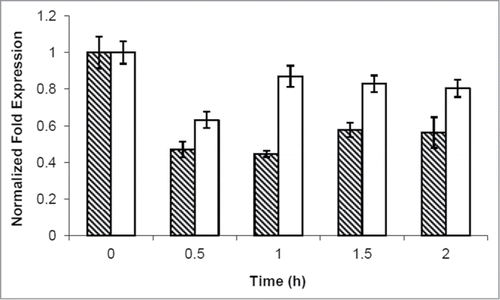
Figure 7. GG104 swi4Δ mutant does not show G1/S transient arrest after PKA activation. Asynchronous cultures of the GG104 swi4Δ growing in YPD medium, GG104 swi4Δ [YCplac111-SWI4S159A], GG104 swi4Δ [YCplac111-SWI4] strains growing exponentially in selective medium were splitted into 2 fractions and 2mM cAMP was added to one of them (▪), while the other was the untreated control (Δ). Samples were harvested every 30 minutes to assay cell number, cell size and budding index.
![Figure 7. GG104 swi4Δ mutant does not show G1/S transient arrest after PKA activation. Asynchronous cultures of the GG104 swi4Δ growing in YPD medium, GG104 swi4Δ [YCplac111-SWI4S159A], GG104 swi4Δ [YCplac111-SWI4] strains growing exponentially in selective medium were splitted into 2 fractions and 2mM cAMP was added to one of them (▪), while the other was the untreated control (Δ). Samples were harvested every 30 minutes to assay cell number, cell size and budding index.](/cms/asset/1d93bfbe-ad5e-4f06-96d6-0cb8b4104f34/kccy_a_1055997_f0007_b.gif)
Table 2 Yeast strains used in this study
