Figures & data
Figure 1. Identification of 3 major SUMOylation sites on TopoIIα-CTD. (A) In vitro SUMOylation was performed for T7-tagged TopoIIα-CTD fragments under the indicated conditions. The reactions were analyzed by immunoblotting with HRP-conjugated anti-T7 antibody. Bracket indicates SUMOylated TopoIIα-CTD (S-CTD), and a bar indicates non-SUMOylated TopoIIα-CTD. (B) TopoIIα-CTD primary sequence. The bold letters indicate the lysines tested for SUMOylation capability. SUMO acceptor lysines were underlined. (C) The 3 underlined lysines in (B) were mutated to arginine (CTD-3KR). Both CTD and CTD-3KR were subjected to SUMOylation assays with the indicated concentrations of enzymes and collected at the indicated time points. S-CTD and CTD are shown with a bracket and bar, respectively. (D) XEEs were immunodepleted using non-specific IgG (Cont.) or anti-TopoIIα antibody (−TopoIIα) and the depletion efficiency was confirmed by immunoblotting the extracts (left 2 lanes, labeled XEE). WT or mutant TopoIIα (3KR) was added to the TopoIIα-depleted extracts. The mitotic chromosomes were analyzed by immunoblotting with anti-TopoIIα. The reaction with dominant negative mutant Ubc9 (dnUbc9) was included as a negative control (+dn). SUMOylated TopoIIα is indicated with a bracket.
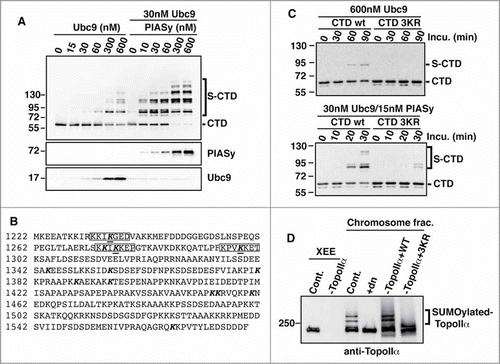
Figure 2. The SUMOylation of TopoIIα-CTD is not required for the SUMOylation-dependent inhibition of decatenation activity. (A) Both unSUMOylated (−) and SUMOylated TopoIIα (+) WT, K660R, 3KR, and 4KR were incubated with kinetoplast DNA (kDNA) for the indicated time. The reactions were resolved in an agarose gel to separate decatenated (bracket) from catenated DNA (arrow). (B, C) The band intensity of catenated kDNA from 3 independent experiments was measured as described in the Materials and methods. The relative amount of remaining catenated kDNA is presented as % catenated kDNA (F), and the inhibition rate by SUMOylation of each reaction was calculated by (% catenated DNA in +SUMO reaction)/(% catenated DNA in -SUMO reaction) in each assay with standard error.
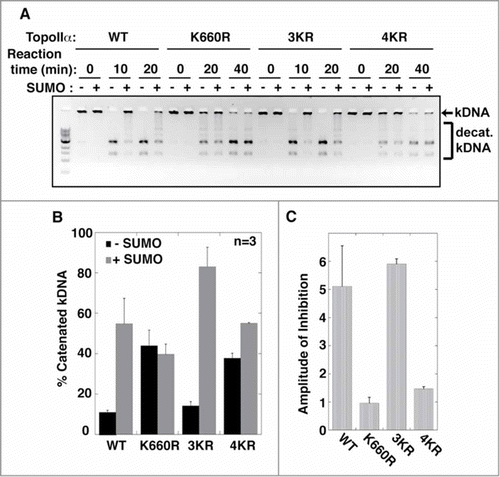
Figure 3. SUMOylation of TopoIIα-CTD promotes protein interactions. (A) Both mock-SUMOylated (CTD) and SUMOylated CTD (S-CTD) were bound to S-agarose beads and incubated with either XEE (+XEE) or buffer (−XEE). Empty S-agarose beads were used as a negative control (indicated as beads). Proteins that bound to the beads were visualized with silver staining. The asterisks indicate bands observed only in the S-CTD-bound beads when incubated with XEE. (B) The pulled-down beads were incubated with the SENP2 catalytic domain (SENP2-CD, indicated with an arrow), and soluble fractions were visualized with silver staining. Digested SUMO2 moiety from the S-CTD is indicated with an arrow. The eluted binding protein of approximately 250 kDa (an asterisk) was identified as Claspin by LC-MS/MS analysis. (C) The SENP2 elution fractions in panel B) were analyzed by immunoblotting with anti-Claspin antibody. The input represents 10% of the XEE used for the pull-down assays. (D) The pulled-down fractions with SUMO chain (GFP-SUMO2x4) were analyzed by immunoblotting using anti-Claspin and anti-PICH antibodies.
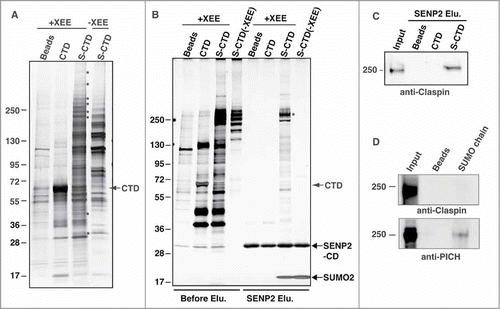
Figure 4. TopoIIα-CTD SUMOylation is required for the robust binding of Claspin to mitotic chromosomes. (A) Mitotic chromosomes were prepared from XEE with (Cont.) or without (+dn) mitotic SUMOylation. The isolated chromosomes were analyzed for the indicated proteins by immunoblotting. SUMOylation of TopoIIα and PARP1, both of which are established mitotic chromosomal SUMOylated proteins, is inhibited by addition of dnUbc9. Molecular weight markers are shown on each side (kDa). (B) Four independent experiments as in A) were performed. The relative amount of chromosome-bound Claspin was normalized with RCC1 and measured as described in the Materials and methods. “Cont.” and “+dn” indicate the conditions with and without SUMOylation, respectively. Standard error is shown with a bar. (C) Endogenous TopoIIα was replaced with either recombinant TopoIIα-WT, K3R or K4R. The efficiency of depletion was confirmed by immunoblotting (left panel, Extracts). Mitotic chromosomes from TopoIIα-replaced XEEs were analyzed by immunoblotting with the indicated antibodies. The SUMOylated form of TopoIIα is indicated by a bracket.
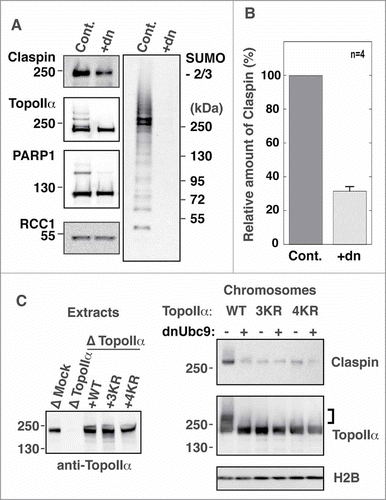
Figure 5. The centromeric localization of Claspin is regulated by mitotic SUMOylation. Mitotic chromosomes prepared from XEEs were subjected to immunofluorescence staining with the indicated antibodies. The right panels are merged images of TopoIIα in red, Claspin in green and DNA in blue. Upper panels) A group of chromosomes. Lower panels) A single pair of sister chromatids. Bars indicate 10 μm.
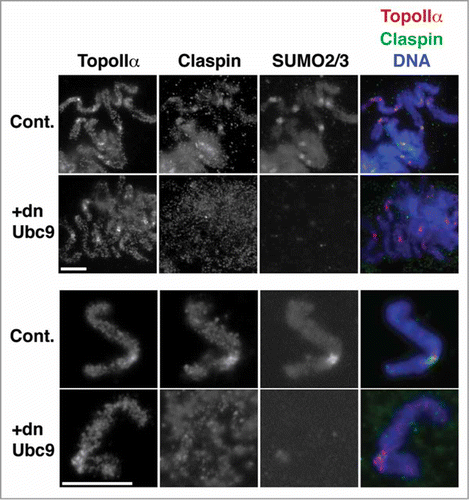
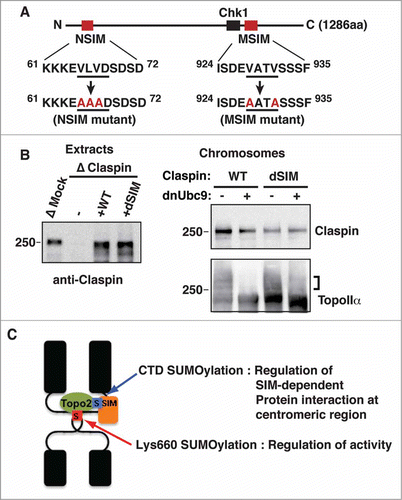
Figure 6. SIMs on Claspin are required for its SUMOylation-dependent binding to mitotic chromosomes. (A) A schematic of the primary structure of Claspin. The SIMs and Chk1 binding domainsCitation42 are indicated as red and black boxes, respectively. The point mutations in each SIM are red. The residue numbers are indicated on the side. (B) Endogenous Claspin was replaced with either recombinant WT or a dSIM mutant by immunodepletion/add-back. The efficiency of depletion and amount of supplemented recombinant proteins were confirmed by immunoblotting (left panel, Extracts). Mitotic chromosomes from Claspin-replaced XEEs were analyzed by immunoblotting with the indicated antibodies. The SUMOylated form of TopoIIα is indicated by a bracket. (C) A proposed model of the dual function of TopoIIα SUMOylation on mitotic centromeres.