Figures & data
Figure 1. Apoptotic activity was analyzed by the terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) assay. (A) Positive TUNEL straining (green) was observed under a fluorescence microscope. (B) Quantitative analysis. The number of apoptotic cells was calculated by averaging the number of positive TUNEL signals. *Significant interstrain differences, р < 0.05; #significant differences with the preceding age, р < 0.05. Bar 50 mkm.
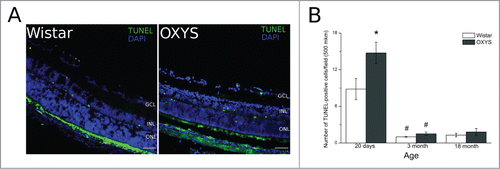
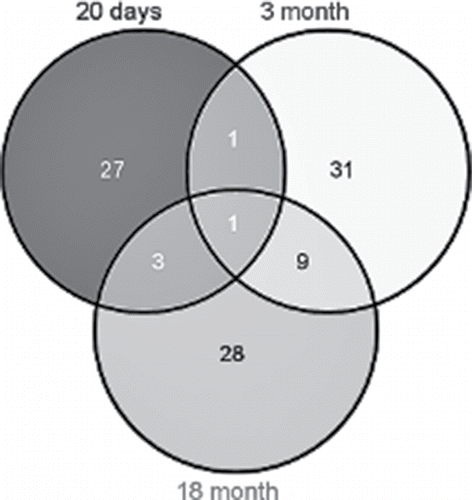
Table 1. Differentially expressed genes (DEGs) with a fold change (FC) cutoff > 2.0 and adjusted p values < 0.05
Figure 2. Statistically significant (p < 0.05) Gene Ontology terms that are related to the genes whose expression is changed in the retina of OXYS rats in comparison with Wistar rats at age 20 d.
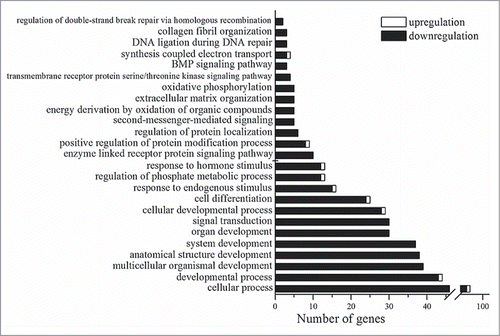
Figure 4. Illustration of the association networks of apoptosis in 20-day-old (A), 3-month-old (B), and 18-month-old rats (C). In each of these networks, black circles denote differentially expressed genes (DEG), whereas the GeneMANIA-predicted genes are shown in gray.
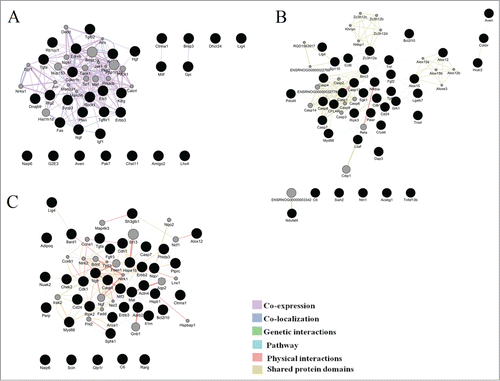
Table 2. Topological parameters of the nodes of association networks related to apoptosis in rats at ages 20 d and 3 and 18 months. The optimal parameters are in boldface
Table 3. Cell death gene ontology (GO) terms for differentially expressed genes (DEGs) and additional related genes according to GeneMANIA analysis for each category (20 days, 3 months, and 18 months)
Figure 5. Clusters in the schemes of gene networks for 20-day-old (A), 3-month-old (B), and 18-month-old rats (C). In each of these networks, black circles denote differentially expressed genes (DEG), whereas the GeneMANIA-predicted genes are shown in gray. The hub genes are yellow
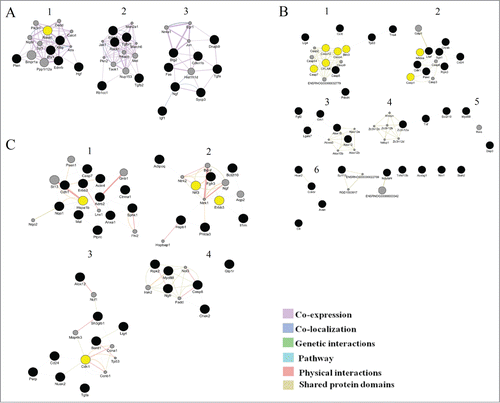
Table 4. Cluster analysis of gene networks for 20-day-old, 3-month-old and 18-month-old rats. Showing clusters with the number of nodes more than 3