Figures & data
Figure 1. Subcellular localization of Rev7. Co-immunostaining images of HeLa cells showing the subcellular localization of Rev7 during metaphase and anaphase stages of the cell cycle with respect to the spindle. During metaphase Rev7 looks to be concentrated around the metaphase plate as detected with a mouse anti-Rev7 antibody, and co-localizes with the spindle as detected with rabbit anti-β-tubulin (top panel). During early anaphase there is an increased Rev7 staining between the anaphase plates (middle panel) and in late anaphase Rev7 staining is reduced and more uniform (bottom panel).
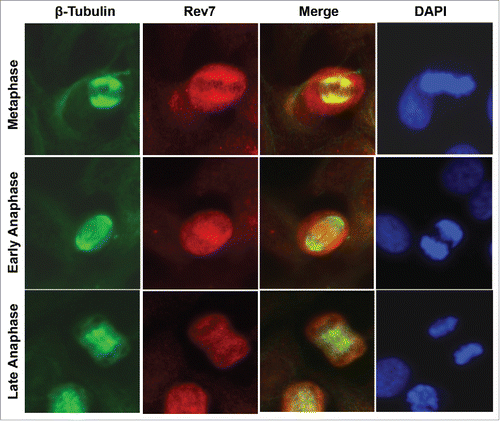
Figure 2. Rev7 depletion causes mitotic arrest in HeLa cells. (A) Representative images showing increased number of round cells (red arrowheads) in siRev7-treated cells after 72 hrs of treatment. (B) Quantitative analysis of the percentage of round cells in mock-, siCTRL- and siRev7-treated cells 72 hrs post-transfection. Results are the average of 3 experiments with approximately 500 cells in each experiment. Round cells were considered to be mitotic cells and the rest to be other phases of the cell cycle. (C) Cell-cycle distribution as determined by flow cytometry after 72 hrs of treatment with indicated siRNAs. siMad2 cells were analyzed after 48 hrs of treatment. (D) Percentage distribution of cells in different phases of the cell cycle averaged from 3 experiments as analyzed from the flow cytometry data in (C). (E) Representative images of DAPI-stained cells with Rev7, RAN or Mad2 depletion showing different types of nuclear abnormalities such as nuclear constrictions (siRAN), nuclear fragmentation (siMad2), interphase bridge and micronuclei formation (siMad2). siRAN images were captured at 36 hrs post-transfection, siMad2 at 48 hrs post-transfection and siRev7 at 72 hrs post-transfection. (F) Quantitative analysis of various nuclear abnormalities observed after the siRNA treatment. I-bridges (Interphase bridges), N-fragmentation (Nuclear fragmentation). Error bars represent standard deviation. *p < 0 .05, **p < 0 .005, ***p < 0 .0005 vs. siCTRL.
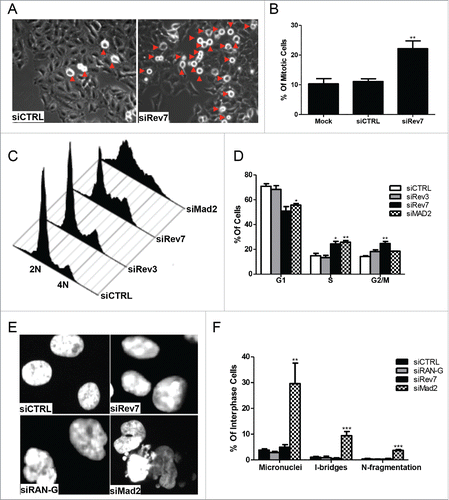
Figure 3. Rev7 and RAN depletion causes abnormal mitotic spindle formation and chromosome misalignment in HeLa cells. (A) Representative images of β-tubulin and pericentrin co-stained cells showing monoastral and multiastral spindles. (B) Quantitative analysis showing the percentage of metaphase cells carrying monoastral and multiastral spindles in 72-h siCTRL- and siRev7-treated samples. (C) Quantitative analysis of the percentage of metaphase cells carrying monoastral and multiastral spindles after 36-h treatment of siRev7 or siRAN. (D) Representative images of β-tubulin-stained cells showing normally-aligned (top panel), mildly-misaligned (middle panel) and severely-misaligned (bottom panel) chromosomes at the metaphase plate. (E) Quantitative analysis of the percentage of metaphase cells carrying misaligned chromosomes after 36 hrs of siRNA treatment. (F) Quantitative analysis of the percentage of metaphase cells carrying misaligned chromosomes after 72 hrs of siRev7 treatment. Error bars represent standard deviation from 3 independent experiments for all figures except B, which were calculated from 6 experiments. *p < 0 .05, **p < 0 .005, ***p < 0 .0005 vs. siCTRL.
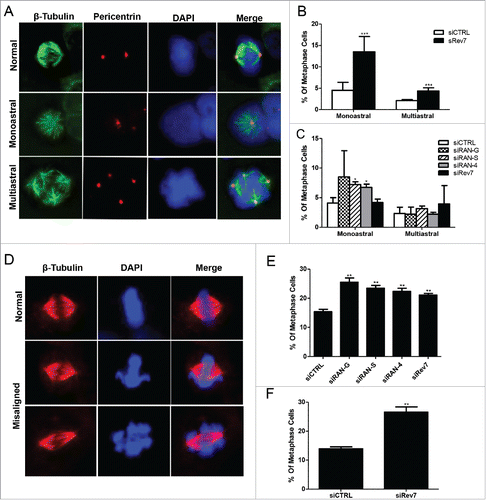
Figure 4. HeLa cells depleted of Rev7 or Mad2 respond differently to nocodazole-induced G2/M cell cycle arrest. (A) Western blots showing partial and complete depletion of Mad2 using different concentrations of siMad2. β-tubulin was used as an internal control. (B) Representative images taken from cells treated with siRev7 or 6.5 nM siMad2 for 48 hrs followed by treatment with 200 ng/ml nocodazole for an additional 24 hrs before taking photographs. (C) Quantitative analysis of the percentage of interphase (flat) and metaphase (round) cells in different treatment groups averaged from 2 experiments with approximately 300 cells in each experiment.
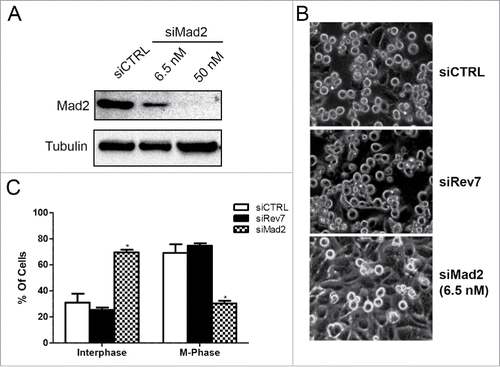
Figure 5. Rev7 depletion induces Mad2 focus formation and Cyclin B1 accumulation in HeLa cells. (A) Representative images of Mad2- and β-tubulin-immunostained cells showing a normal metaphase cell with no Mad2 foci (left panel) and a misaligned metaphase cell with Mad2 foci and an abnormal spindle (right panel). (B) Quantitative analysis of metaphase cells positive for Mad2 foci 72 hrs after siRev7 treatment. (C) Western blots showing increased levels of Cyclin B1 after Rev7 depletion at 0 hr post-release from RO-3306 block and delayed degradation at 45 and 96 minutes post-release when compared to siCTRL treated cells. Mad2 levels remained unchanged. Actin was used as an internal control. Error bars represent standard deviation from 3 experiments. **p < 0.005 vs. siCTRL.
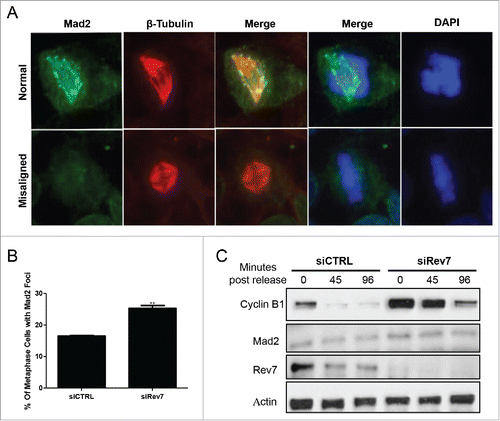
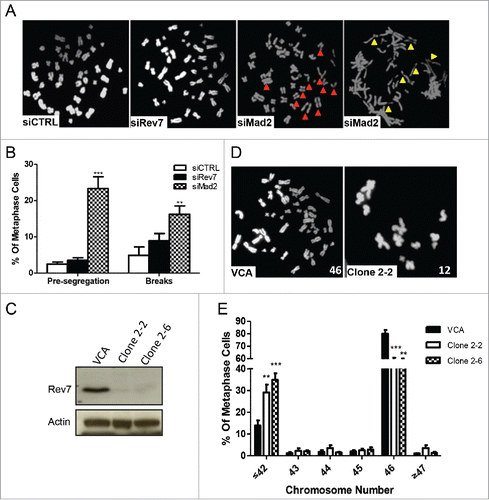
Figure 6. Mad2 depletion causes premature segregation and DNA damage whereas long-term depletion of Rev7 causes aneuploidy. (A) Representative images of HCT116 metaphase spread showing a normal metaphase in siCTRL- and siRev7-treated cells but premature sister chromatid segregation (red arrow heads) and chromosome breaks (yellow arrow heads) in siMad2-treated cells 48 hrs post-transfection. (B) Quantitative analysis of premature segregation (Pre-segregation) and chromosomal breaks in images taken from (A). Error bars represent standard deviations from 3 experiments. **p < 0 .005, ***p<0.0005 vs. siCTRL in respective groups. (C) Western blots showing stable depletion of Rev7 in 2 selected clones of MSU1.1 cells. (D) Representative metaphase spreads from vector control MSU1.1 cells (VCA) and Rev7-depleted MSU1.1 cell clone 2–2 showing aneuploidy. (E) Quantitative analysis of metaphase spreads from Rev7-depleted MSU1.1 cell clones 2–2 and 2–6 showing the percentage of metaphase cells with ≤42 chromosomes. **p < 0 .005, ***p < 0 .0005 vs. VCA.