Figures & data
Figure 1. BRMS1 is a novel CDK substrate. (A) Domain structure of BRMS1, which contains imperfect Leucine zippers, coiled-coil domains, nuclear localization sequences and a potential CDK phosphorylation site, located at serine 237. (B) BRMS1 is phosphorylated in cells by CDKs. Left panel, HEK-293T cells transfected with pCMV-Tag2A vector (Lane 1) or pCMV-Tag2A-BRMS1 (Lane 2 and 3), were 32P-labeled in the absence (Lanes 1 and 3) or presence (Lane 2) of 50 μM Roscotivine. Immunoprecipitated FLAG-tagged BRMS1 was separated by SDS-PAGE and transferred onto nitrocellulose. BRMS1 phosphorylation was detected by autoradiography (top panel). Western blotting was performed with an anti-FLAG antibody to demonstrate equal levels of BRMS1 in lanes 2 and 3 (bottom panel). Right panel: Phosphorylated BRMS1 purified from transfected HEK-293T cells was subjected to phosphoamino acid analysis. Phosphoamino acids were visualised by ninhydrin staining and autoradiography. Arrows indicate the positions of phosphoserine (P-Ser), phosphothreonine (P-Thr) and phoshotyrosine (P-Tyr). (C) In vitro phosphorylation of BRMS1 by Cyclin A/CDK2. Left panel: purified His6-tagged wild-type BRMS1 or BRMS1 S237A were incubated with or without Cyclin A/CDK2 in the presence of [γ-32P] ATP. Following phosphorylation, samples were separated on SDS-PAGE, stained with Coomassie brilliant blue (bottom panel) and autoradiographed (top panel). Right panel: purified His6-tagged BRMS1 phosphorylated in vitro by Cyclin A/CDK2 in the presence of [γ-32P] ATP was subjected to phosphoamino acid analysis. Phosphoamino acids were visualized by ninhydrin staining and autoradiography. Arrows indicate the positions of phosphoserine (P-Ser), phosphotheorine (P-Thr) and phosphotyrosine (P-Tyr). (D) BRMS1 is phosphorylated on Ser 237. In vitro or in vivo phosphorylated BRMS1 (from B and C) was separated by SDS-PAGE, excised and subjected to tryptic digestion. Phosphopeptides were then purified on TiO2 beads and analyzed by LC/MS. Tandem MS/MS mass spectra of phosphorylated serine 237 (pS237) from His6-BRMS1 phosphopeptide 234–241 (AAVpSPQKR), parent ion: 468.8 m/z 2+ (indicated with an arrow), following phosphorylation in vitro by CyclinA/CDK2 (top panel), or following immunoprecipitation of FLAG-tagged BRMS1 from BT-549 cells (bottom panel).
![Figure 1. BRMS1 is a novel CDK substrate. (A) Domain structure of BRMS1, which contains imperfect Leucine zippers, coiled-coil domains, nuclear localization sequences and a potential CDK phosphorylation site, located at serine 237. (B) BRMS1 is phosphorylated in cells by CDKs. Left panel, HEK-293T cells transfected with pCMV-Tag2A vector (Lane 1) or pCMV-Tag2A-BRMS1 (Lane 2 and 3), were 32P-labeled in the absence (Lanes 1 and 3) or presence (Lane 2) of 50 μM Roscotivine. Immunoprecipitated FLAG-tagged BRMS1 was separated by SDS-PAGE and transferred onto nitrocellulose. BRMS1 phosphorylation was detected by autoradiography (top panel). Western blotting was performed with an anti-FLAG antibody to demonstrate equal levels of BRMS1 in lanes 2 and 3 (bottom panel). Right panel: Phosphorylated BRMS1 purified from transfected HEK-293T cells was subjected to phosphoamino acid analysis. Phosphoamino acids were visualised by ninhydrin staining and autoradiography. Arrows indicate the positions of phosphoserine (P-Ser), phosphothreonine (P-Thr) and phoshotyrosine (P-Tyr). (C) In vitro phosphorylation of BRMS1 by Cyclin A/CDK2. Left panel: purified His6-tagged wild-type BRMS1 or BRMS1 S237A were incubated with or without Cyclin A/CDK2 in the presence of [γ-32P] ATP. Following phosphorylation, samples were separated on SDS-PAGE, stained with Coomassie brilliant blue (bottom panel) and autoradiographed (top panel). Right panel: purified His6-tagged BRMS1 phosphorylated in vitro by Cyclin A/CDK2 in the presence of [γ-32P] ATP was subjected to phosphoamino acid analysis. Phosphoamino acids were visualized by ninhydrin staining and autoradiography. Arrows indicate the positions of phosphoserine (P-Ser), phosphotheorine (P-Thr) and phosphotyrosine (P-Tyr). (D) BRMS1 is phosphorylated on Ser 237. In vitro or in vivo phosphorylated BRMS1 (from B and C) was separated by SDS-PAGE, excised and subjected to tryptic digestion. Phosphopeptides were then purified on TiO2 beads and analyzed by LC/MS. Tandem MS/MS mass spectra of phosphorylated serine 237 (pS237) from His6-BRMS1 phosphopeptide 234–241 (AAVpSPQKR), parent ion: 468.8 m/z 2+ (indicated with an arrow), following phosphorylation in vitro by CyclinA/CDK2 (top panel), or following immunoprecipitation of FLAG-tagged BRMS1 from BT-549 cells (bottom panel).](/cms/asset/80cc1bcb-5cc4-4f98-8c6c-3201eb9b15b0/kccy_a_1121328_f0001_oc.gif)
Figure 2. Phosphorylation of BRMS1 on serine 237 does not affect cell cycle progression or proliferation. (A) Western blot representing stable MDA-MB-231 cells expressing (Lane 1) Vector, (Lane 2) BRMS1-WT, (Lane 3) BRMS1-S237A and (Lane 4) BRMS1-S237D. (B, left histogram) Effect of BRMS1-S 237 on S-phase. MDA-MB-231 cells infected with the pLenti-DEST6/V5 (Vector) or expressing BRMS1-WT, BRMS1-S237A or BRMS1-S237D were assessed for the percentage of cells in S phase. Cells were labeled with BrdU, fixed with 70% ethanol, stained with FITC-conjugated anti-BrdU antibody and analyzed by FACS on a Fortessa cell analyzer. Gating was done based on positive staining for BrdU (left panel). The percentage of cells in the S phase is represented in the histogram; Vector (57.8 %), BRMS1-WT (68.7 %), BRMS1-S237A (69.2 %), BRMS1-S237D (65.6 %). One-Way ANOVA with Post-Tukey test were carried out for analysis. Error bars represent ±SEM from 3 biological samples (B, right histogram) Effect of BRMS1-S 237 on M-phase. To measure the percentage of cells in the mitotic phase, cells were subjected to double staining with AlexaFluro 647-conjugated anti-phosphohistone H3 antibody and Propidium Iodide followed by analysis by FACS on a Fortessa cell analyzer. Cells with double the DNA content that positively stained for phosphohistone H3 were gated (left panel). Quantification was performed using One-Way ANOVA with Post-Tukey test. The percentage of cells in mitosis is represented in the histogram; Vector (2.3 %), BRMS1-WT (1.9 %), BRMS1-S237A (2.2 %), BRMS1-S237D (2.1 %). Error bars represent ±SEM from 3 biological samples. (C) Effect of BRMS1-S 237 on cell proliferation. Cells were plated in 96-well plates and their rate of proliferation was quantified by MTS assay over 7 d. Error bars represent ±SEM from 3 biological samples. (D) Effect of BRMS1-S 237 on colony formation. 3000 cells were seeded onto 6 cm plates and left to grow for 7 d. Colonies were fixed and stained with Crystal Violet (left panel) and the number of colonies was determined using ImageJ (Right panel). Error bars represent ±SEM of 3 biological samples.
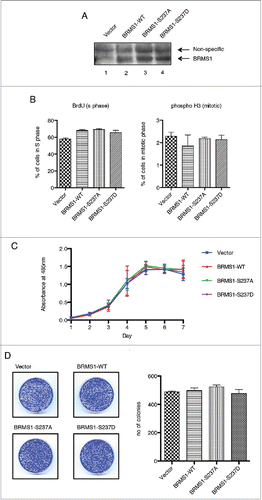
Figure 3. Phosphorylation of BRMS1 on serine 237 regulates cell migration. (A) Scratch-induced migration assay. 90 % confluent MDA-MB-293 cells were scratched with pipette tips. Phase contrast microscope images were taken at 0 and 24 hours post-wound induction from 3 different experiments and the size of the wounds was measured across 5 points, as indicated, and the averages were calculated. The percentage of the initial wound distance covered by the cells after 24h is represented on the histogram (Bottom panel). Error bars represent ±SEM from 3 biological samples. (B) Chemoattractant-induced transwell migration assays. 5 × 104 MDA-MB-231 cells resuspended in 1% FBS supplemented-media were placed in an upper transwell chamber. Cells were then allowed to migrate toward 10% FBS supplemented-media located in the lower chamber for 16 h.Citation67 Following Crystal-Violet staining, non-migratory cells were removed from the inside of the chamber by cotton tips. The stain extracted from the migrating cells was measured at OD560nm using a plate reader. Results were represented as relative fold change compared to control and error bars represent ±SEM from 3 biological samples.
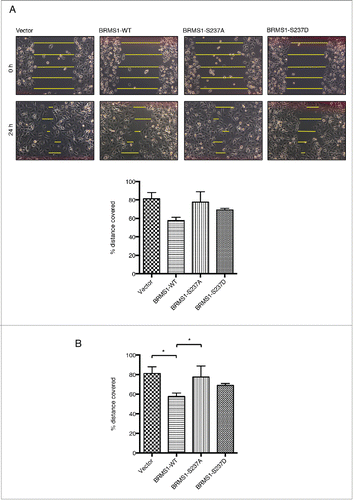
Figure 4. Phosphorylation of BRMS1 on S237 does not affect its association with RBP1 or the mSin3/HDAC complex. (A) Co-immunoprecipitation of RBP1 and BRMS1 mutants. HEK-293T cells were co-transfected with constructs expressing FLAG-tagged RBP1-WT, RBP1-S864A/S1007A or RBP1-S864D/S1007D and Myc-tagged BRMS1-WT, BRMS1-S237A or BRMS1-S237D. Myc-tagged BRMS1 was immunoprecipitated with anti-Myc affinity beads and the co-immunoprecipitated FLAG-tagged RBP1 was detected by Western blotting. Lane 1 represents the control, where the level of non-specific RBP1 binding is assessed on the resin from cells not expressing ectopic BRMS1. Lanes 2–10 represent the levels of the co-immunoprecipiated BRMS1 and RBP1, wild-type or phospho-mutants, as indicated. (B) Co-immunoprecipitation of BRMS1 mutants and mSin3, HDAC1 and SAP30.BT-549 cells were transfected with constructs expressing FLAG-tagged BRMS1-WT (lane 2–3), BRMS1-S237A (lane 4) or BRMS1-S237D (lane 5). 50 μM of the CDK1/2 inhibitor Roscovitine was added to the cells 4 hours prior to lysis, to inhibit the in vivo phosphorylation of BRMS1-WT (Lane 3). BRMS1 was immunoprecipitated from cell lysates with anti- FLAG M2 affinity beads and co-immunoprecipitated mSin3, HDAC1 and SAP30 detected by Western blotting. Lane 1 represents immunoprecipitation from lysates of cells transfected with empty vector.
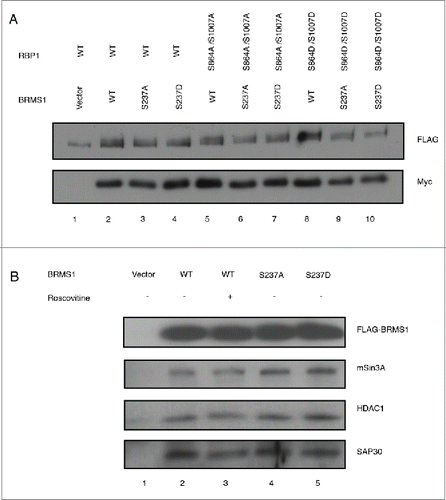
Figure 5. Phosphorylation of BRMS1 serine 237 does not regulate transcriptional repression. (A) Transcriptional regulation of BRMS1-S237. Plasmid expressing Gal4-wild-type-BRMS1, Gal4-BRMS1-S237A or Gal4-BRMS1-S237D were co-transfected into BT-549 cells together with the pGL2-(Gal4)5 TK and pRL-CMV luciferase reporter plasmids. Luminescence of the cell lysates were measured using a plate reader. Firefly luciferase luminescence was normalized against Renilla luciferase luminescence. The graph represents the mean luminescence ±SEM from 3 independent experiments. The western blot (lower panel) demonstrates the levels of BRMS1 expression in these cells. (B) Transcriptional regulation of OPN by BRMS1-S237. qRT-PCR of mRNA from MDA-MB-231 cells expressing BRMS1-WT, BRMS1-S237A, BRMS1-S237D. mRNA levels of, ectopic BRMS1 and OPN were quantified and plotted as relative fold change compared to vector control cells. Error bars represent ±SEM from 3 independent experiments.
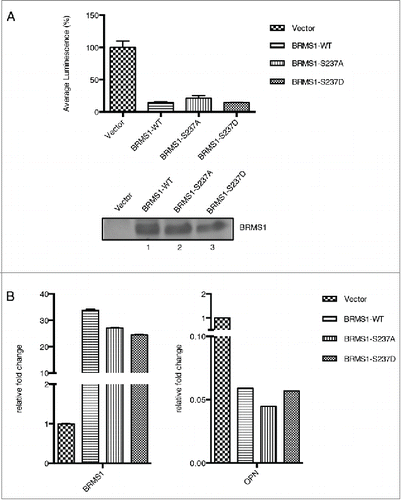
Figure 6. Phosphorylation of on serine 237 does not affect subcellular localization. Subcellular localization of GFP-tagged BRMS1-WT, BRMS1-S237A or BRMS1-S237D in BT-549 cells. Transfected cells were plated onto cover slips and nuclear staining (Hoechst) and GFP-BRMS1 localization were visualized using am Olympus BX-51 fluorescence microscope. Fluorescence intensities within the cytoplasm and nucleus were expressed as percentages of the total fluorescence in the cell. The total area of each cell was determined based on Differential Interference Contrast (DIC) images while nuclei area was based on the Hoechst stain. Cytoplasmic to nucleus ratios were calculated as previously described [32]. Error bars represent ±SEM of 3 independent experiments.
![Figure 6. Phosphorylation of on serine 237 does not affect subcellular localization. Subcellular localization of GFP-tagged BRMS1-WT, BRMS1-S237A or BRMS1-S237D in BT-549 cells. Transfected cells were plated onto cover slips and nuclear staining (Hoechst) and GFP-BRMS1 localization were visualized using am Olympus BX-51 fluorescence microscope. Fluorescence intensities within the cytoplasm and nucleus were expressed as percentages of the total fluorescence in the cell. The total area of each cell was determined based on Differential Interference Contrast (DIC) images while nuclei area was based on the Hoechst stain. Cytoplasmic to nucleus ratios were calculated as previously described [32]. Error bars represent ±SEM of 3 independent experiments.](/cms/asset/7e07f272-985c-4f42-a2f8-30f8cd9e274f/kccy_a_1121328_f0006_oc.gif)
