Figures & data
Figure 1. Foxo1 and Foxo3 follow similar expression patterns among HSPC populations. (A) (Top) Hierarchy of HSPC populations examined, with LSK cells at the top encompassing HSCs and multipotent progenitors, followed by CMPs, which give rise to GMPs and MEPs, the myeloid and erythroid lineage specific progenitors, respectively. (Bottom) Sorting scheme of HSPC populations: LSK (LIN- Sca1+ cKit+), CMP (LIN- IL7Rα- cKIT+ CD34+ Fcc3-), GMP ((LIN- IL7Rα- cKIT+ CD34+ Fcc3+) MEP ((LIN- IL7Rα- cKIT+ CD34- Fcc3-) (B) qRT-PCR analysis of cDNA generated from 3 independent experiments each containing a pool of 3 mice from each genotype. Absolute Cts (cycle which SYBR green fluorescence is detected over set threshold during exponential expansion of Foxo1 or Foxo3 transcripts) were normalized to B-actin levels as a control for total cDNA abundance. B-actin normalized expression levels comparing transcript abundance of Foxo1 and Foxo3 within each population are displayed. (C) B-actin normalized expression levels of Foxo1 and Foxo3 were further normalized to expression within the LSK population (ΔΔCt Method) to examine how Foxo1 and Foxo3 individually differ over HSPC populations. (Bars represent averages of 3 independent experiments. Results are mean ± s.e.m (n=3) *p < 0.5, **p < 0.01 2-way ANOVA).
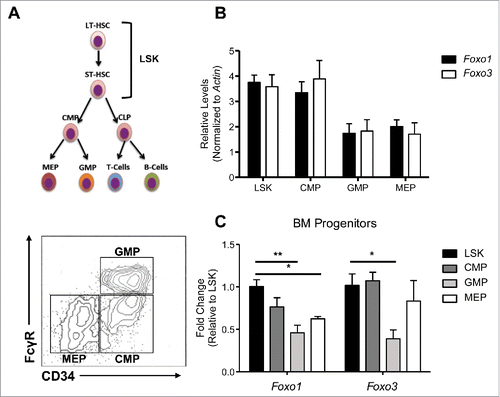
Figure 2. FOXO3 is significantly more active than FOXO1 based on nuclear localization within each HSPC population. (A) Using the same sorting strategy as in 1A (bottom), each population was cytospun and fixed onto slides for immunofluorescence analysis of FOXO1 or FOXO3 subcellular localization. Representative immunofluorescence images displaying FOXO1, FOXO3, or pAKT in red and counterstained with DAPI to delimit the nucleus, shown in blue. Columns separate each population and rows separate protein examined. (B) Thirty to 40 cells per population were quantified over 3 independent experiments. Nuclear localization was determined by proportion of total FOXO1 or FOXO3 fluorescence within area delimited by DAPI using ImageJ and MATLAB to measure fluorescence values and calculate proportions. With 0 representing complete cytoplasmic FOXO and 1 representing complete nuclear FOXO (Bars represent mean ± s.e.m. (n > 30) from 3 independent experiments with BM cells from a pool of 3 mice per experiment. *p < 0.5, **p < 0.01, ***p < 0.001 students t-test).
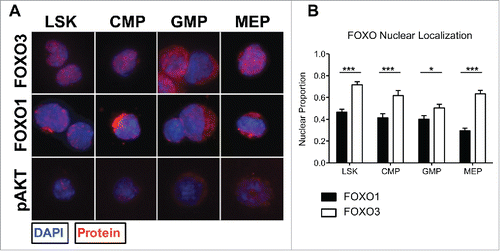
Figure 3. HSPC FOXO1 and FOXO3 nuclear localization is insensitive to AKT inhibition by LY294002 (LY). (A-B) Total BM cells were isolated from 4 mice per genotype and stained with antibodies specific to cell surface markers discussed in (bottom) to distinguish HSPC populations. Cells were then treated with DMSO or 10uM LY294002 for 30 min followed by fixation and permeablization. Subsequently, flow cytometry analysis of pAKT (Ser 473), total FOXO1, total FOXO3, pFOXO1 (Ser 253), and pFOXO3 (Ser256) levels with specific fluorescence conjugated antibodies. Graphs represent abundance of protein denoted by the geometric mean of antibody fluorescence bound to protein. (Error bars indicate s.e.m. *p < 0.5, **p < 0.01 paired t-test) (C) BM cells treated with DMSO and LY294002 were also sorted as in (bottom) and cytospun onto slides for immunofluorescence analysis of FOXO1 and FOXO3. Thirty-40 cells per population were analyzed over 3 independent experiments. Graph represents quantification (same method as described in ) of the fold change in nuclear localization relative to DMSO control of either FOXO1 or FOXO3. (Bars represent mean ± s.e.m. (n > 30)).
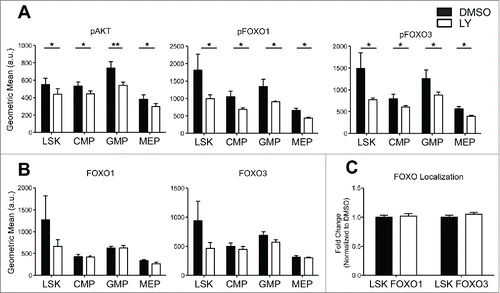
Figure 4. FOXO3 localization is sensitized to modulation by AKT in the absence of SIRT1 in LSK cells. A,B. Sorted LSK cells from WT and Sirt1−/− mice were cytospun onto slides for immunofluorescence analysis for either FOXO1 (A) or FOXO3 (B). LSK cells were then treated with DMSO or LY294002 (LY) as in previous experiments. Graph represents quantification of fold change in nuclear localization relative to DMSO control of either FOXO1 or FOXO3. (Bars represent mean ± s.e.m. (n > 40), from at least 2 independent experiments. *p < .05, students t-test).
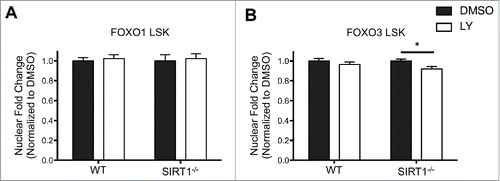
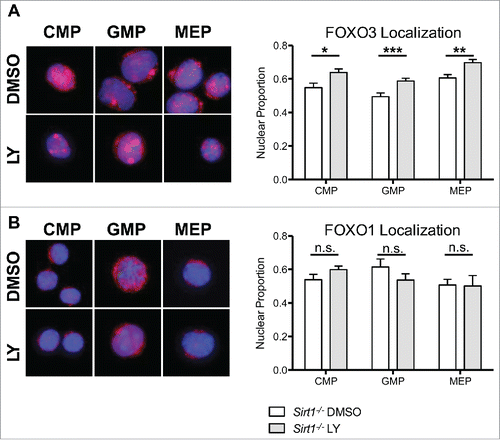
Figure 5. Increased FOXO3, but not FOXO1, nuclear localization induced by AKT inhibition in Sirt1−/− hematopoietic progenitor cells. (A) Sorted CMP, GMP, and MEP cells from BM cells of Sirt1−/− mice were treated with DMSO or LY294002 (LY) as in previous experiments. (Left) Representative images of CMP, GMP, MEP cells treated with DMSO or LY294002, with DAPI delimiting the nucleus in blue, and FOXO3 in red. (Right) Graphs displays quantification of the proportion of FOXO3 within the nucleus. (B) Same as in (A) but with FOXO1. (Bars represent mean ± s.e.m. (n > 20), from at least 2 independent experiments. *p < .05, **p < .01, ***p < .001, students t-test). Note the similar fraction of nuclear FOXO1 and FOXO3 in Sirt1−/− CMP.