Figures & data
Figure 1. Nuclear localization of Plk1 in human pancreas cells. A, HPDE6, Pa03C, Panc1 and Panc10.05 were treated with or without 200 ng/ml nocodazole (Noc) for 12 h and harvested for IB. B, Pa03C, Panc10.05 and HPDE6 cells were treated with 0.3 mM mimosine (Mimo) for 20h, 4 mM hydroxyurea (HU) for 24 h, or 200 ng/ml nocodazole for 12 h to block cells at G1, S, or M phase, respectively. Cytoplasmic and nuclear fractions were isolated and analyzed by Western blot. C, Pa03C or Panc10.05 cells randomly growing on coverslips were stained with a Plk1 antibody and analyzed by IF microscopy. DNA is visualized with DAPI staining.
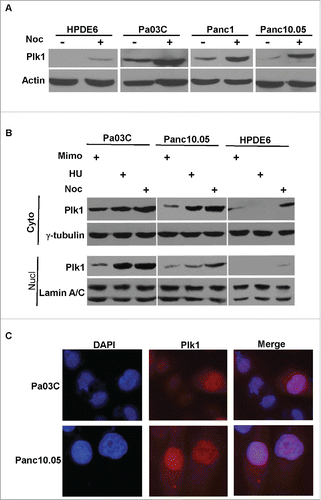
Figure 2. Plk1 depletion in human pancreatic cancer cells. A, Pa03C cells were infected with lentivirus to deplete Plk1. 36h after lentivirus infection, puromycin was added for an additional 36 h to select for infection-positive cells. After floating cells were removed, the remaining attached cells were treated with 200 ng/ml nocodazole for different times as indicated and harvested for IB. B, Panc1 cells were co-transfected with pBS/U6-Plk1 and pBabe-puro at a ratio of 8:1. At 1 d post-transfection, puromycin was added for an additional 36 h to select for transfection-positive cells. After floating cells were removed, the remaining attached cells were harvested for IB. C, Randomly growing Panc1 cells were depleted of Plk1 using the protocol described in B, cells were treated with or without 200 ng/ml nocodazole and harvested at different time points for FACS.
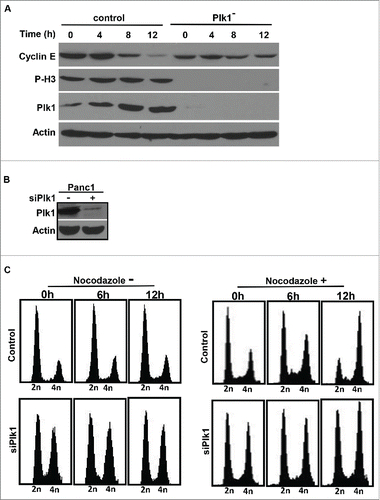
Figure 3. Effects of inhibition of Plk1 on DNA replication. A, FACS analysis of cell cycle progression in Pa03C cells. After Pa03C cells were synchronized with a double thymidine block (DTB; 16 h treatment with thymidine, 8 h release, and a second thymidine block for 16 h), synchronized cells were released into medium in the absence or presence of GSK461364A (25nM or 100nM) and harvested at different time points for FACS. GSK461364A was added into the medium1 h prior to release. B, Representative images of BrdU labeling in control and Plk1-inhibited cells. Pa03C cells were synchronized with the DTB, released into medium with or without GSK461364A (added 1 h prior to release) for 2h or 5h, labeled with BrdU for 30 min, and stained with anti-BrdU antibodies. DNA was stained with DAPI. Scale bar, 100 µm. C, Quantification of BrdU-positive cells in B.
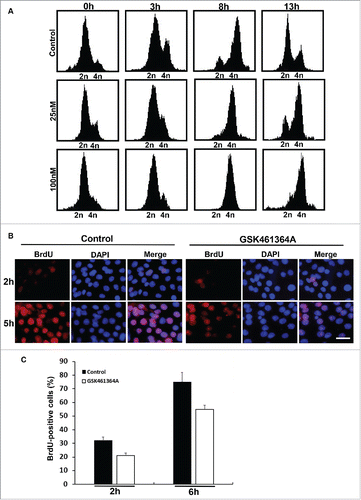
Figure 4. GSK461463A and gemcitabine synergistically inhibit growth of Panc1-derived orthotopic xenograft tumors. A, Panc-1 cells were treated with GSK461364 (10 nmol/L), gemcitabine (100 nmol/L), or both for 72 h, followed by Western blot. B, The combination index of GSK461364A, gemcitabine in Panc1 cells. C, Panc1-derived orthotopic xenograft tumors that had been treated with GSK461364A (12 mg/kg), gemcitabine (40 mg/kg) or a combination of both drugs for 6 weeks. NSG mice were inoculated with Panc1 cells (5 × 105) for 2 weeks, intravenously injected with GSK461364A, gemcitabine or a combination of both drugs. D, Quantification of tumor weight in C. Data are expressed as mean ± SEM, n = 6; **, p < 0.01.
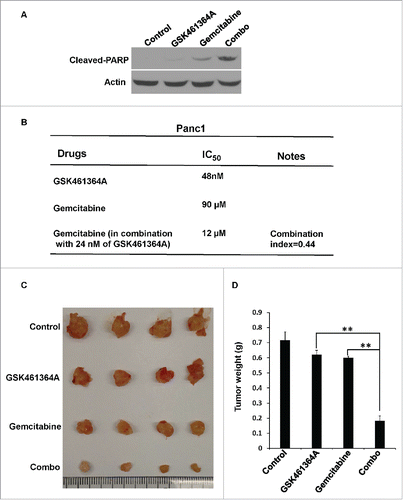
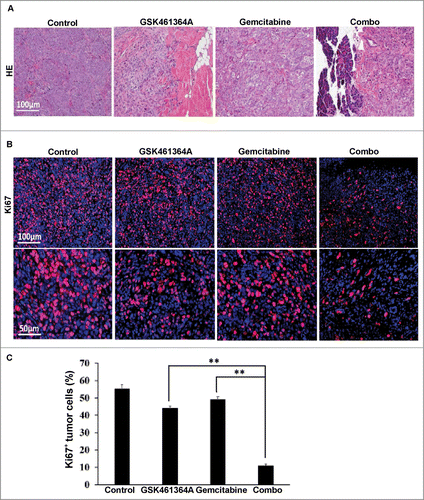
Figure 5. Histological analysis of Panc1-derived orthotopic xenograft tumors. A, Representative images of H&E on formaldehyde-fixed, paraffin-embedded Panc1-derived orthotopic xenograft tumors sections from different treatment groups. B, Representative images of IFC staining for Ki67 on formaldehyde-fixed, paraffin-embedded Panc1-derived orthotopic xenograft tumors sections from different treatment groups. C, Microscopic quantification of Ki67 as percentages of Ki67-positive cells to total numbers of cells. Multiple tumor sections were calculated (mean ± SEM; n = 4). **, p < 0.01.