Figures & data
Figure 1. Chronic acidic microenvironment inhibits proliferation. (A) Cell density and morphology of HCT15 cells (left), HCT116 (middle) and SW480 (right) at pH6.5 were examined under microscopy (×20). HCT15 and HCT116 cells growing in acidic medium were more scattered and elongated. SW480 showed little morphological change except a reduction in cell density. (B) Acidic microenvironment reduced proliferation of CRC cells. Proliferation of HCT15 (top) and HCT116 (bottom) cells cultured at low pH (6.5) for 72 h or for more than 3 months (CRC-AA) were determined by EdU incorporation. Red, EdU; blue, DAPI. (C) Cell cycle distribution of CRC cells growing in acidic medium for various durations. (D) Apoptotic levels of CRC cells adapted to acidic pH (HCT15 and HCT116 cells) and SW480 cells. *, p < 0.05; **, p < 0.01.
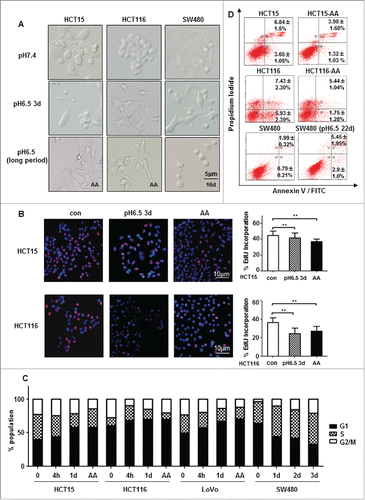
Figure 2. ROS is reduced in colon cancer cells acclimated to acidic microenvironment. (A) DCF fluorescent intensity of HCT15, HCT116, LoVo and SW480 cells exposed to acidic medium for 4 h. (B) DCF fluorescent intensity of HCT15 cells, HCT116 cells and LoVo cells growing acidic medium for more than 3 months. *, p < 0.05; **, p < 0.01.
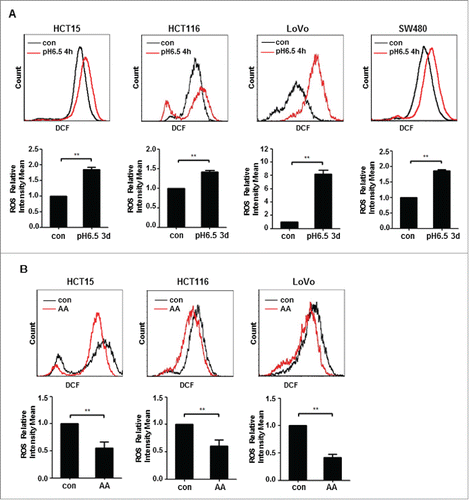
Figure 3. Antioxidant GSH is critical for survival of acidosis-acclimated cancer cells. (A) GSH level as well as GSH/GSSG ratio is higher in HCT15-AA than in their parental cells. The GSH and GSH/GSSG ratio were measured by a GSH and GSSG assay kit. (B) Inhibition of colony formation by BSO in HCT15-AA and their parental cells. Cultured cells were exposed to BSO (10μM) for 12d to allow for colony formation. (C) Induction of ROS by BSO. ROS fluorescence intensity of HCT15 cells treated with BSO (500μM, 12h) was determined by flow cytometry. (D) Inhibition of proliferation of CRC-AA cells and their parental cells by DEM. CRC cells were treated with different concentrations of DEM (0, 0.1, 1, 50μM) for 48 h. Cell viability was measured by MTT assay. *, p < 0.05; **, p < 0.01.
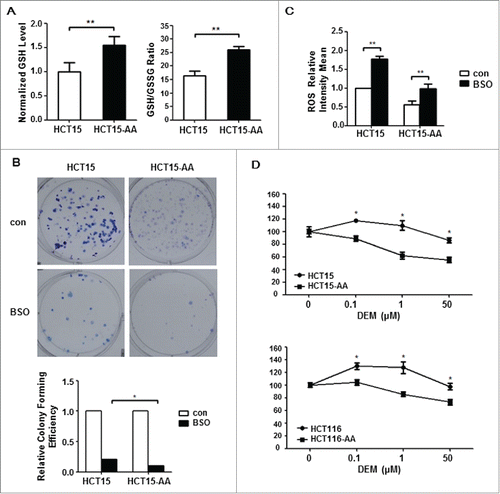
Figure 4. Expression levels of genes involved in GSH metabolism. (A) A diagram showing the key factors involved in GSH metabolism. (B) Relative expressions of representative redox genes at mRNA level in CRC-AA and their parental cells, as measured by quantitative real-time PCR. (C) Expression levels of redox proteins in CRC-AA and their parental cells measured by Western blotting. (D) Flow cytometric analysis of CD44 expression levels in CRC-AA and their parental cells. *, p < 0.05; **, p < 0.01.
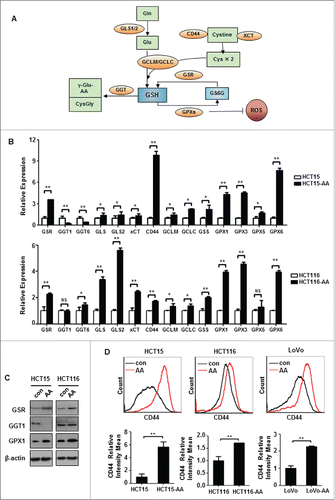
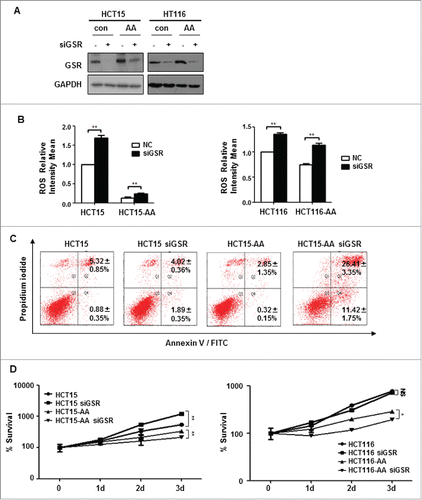
Figure 5. GSR knockdown compromises the survival of CRC-AA cells. (A) Knockdown of GSR by siRNA in HCT15 and HCT116 cells. RNAi efficiency was determined by Western blotting, GAPDH was used as a loading control. (B) Increase of ROS levels in CRC cells by GSR siRNA. (C) Apoptosis was greatly increased in CRC-AA cells when transfected with GSR siRNA. (D) Viability of CRC-AA cells was reduced when transfected with GSR siRNA, as determined by MTT assay. *, p < 0.05; **, p < 0.01.