Figures & data
Figure 1. Ring1b and Med12 target key developmental genes in mouse ESCs. (A) Venn diagram and heatmap of Med12, Ring1b and Cdk8 target genes in mESCs. For the Cdk8 dataset (GSE44288) we performed peak calling with a q value of 1e-5 and annotations with the closest gene promoter (+/−5 kb around the TSS). For the other data sets the qualitative occupancy data were downloaded from the supplementary material of the respective publications. (B) Venn diagram of common Ring1b-Med12 target genes (Ring1b-Med12 only-without Cdk8-, 1670 genes) that overlap with Med1 target genes. (C) GO-enriched terms of Med12-Ring1b and (D) Med12-Cdk8 target genes in mESCs. FDR represents the corrected P-value. (E-F) Med12 but not Cdk8 localize with Cbx7-PRC1 at promoters of repressed differentiation genes in mESCs. Med12 and Cdk8, but not PRC1/PRC2, localize at promoters of expressed genes in mESCs. Screenshots from ChIP-seq profiles of Med1, Med12, Cdk8, Ring1b, Cbx7, RYBP and Suz12 for selected genes. The expression levels of the selected genes (Msx2 and Nanog) are shown. (G) Density distributions of Med12+Ring1b (red) and Med12+Cdk8 (blue) target genes expression. The majority of Med12+Cdk8 targets are expressed at high levels in mESCs whereas the Med12+Ring1b targets have low or no expression. The y-axis shows the density (probability). We omitted this axis from the graph as we only intended to focus on the shift in the x-axis.
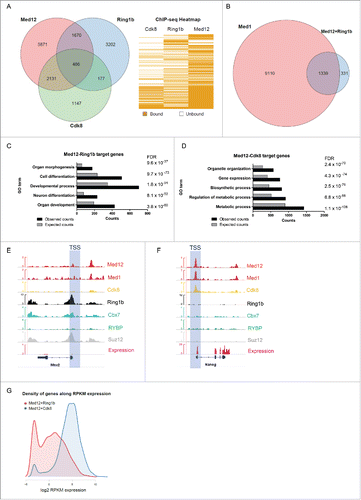
Figure 2. Interdependency of Ring1b and Med12 recruitment to chromatin. (A) Ring1b regulates the recruitment of subunits of the kinase module of Mediator to chromatin. Pluripotent and early differentiating mESCs (shNMC, shMed12 and shRing1b) were fractionated and the chromatin fractions were analyzed employing the respective antibodies. The arrowhead shows the corresponding band for Cdk8. The relative intensities of the protein levels were calculated as fold change to 0 timepoint shNMC samples +/− SEM, N=4, using the ponceau intensities as reference. (B) Ring1b controls the association of Med12 with the core Mediator protein Med1. Endogenous immunoprecipitations (IPs) with Med12 antibodies and protein extracts from control (shNMC) and Ring1b (shRing1b) knockdown mESCs. The precipitated material was analyzed by western blotting with the indicated antibodies. Inputs represent 7% of the material used for each IP. (C) Endogenous IPs with Ring1b antibodies and nuclear extracts from mESCs. The precipitated material was analyzed by western blotting with the indicated antibodies. Inputs represent 7% of the material used for each IP. (D) Ring1b regulates the recruitment of Med12 to promoters of Med12+Ring1b and Med12+Cdk8 target genes in mESCs. Chromatin immunoprecipitations (ChIPs) using chromatin from control (shNMC), Med12 (shMed12) and Ring1b (shRing1b) knockdown mESCs followed by qPCRs on selected promoters. The enrichments are represented as percentage (%) of recovery over input and correspond to the average of 3 independent experiments +/− SEM *** P-value ≤ 0.0003, ** P-value = 0.005, *P-value ≤ 0.03 as calculated by 2-tailed unpaired t-test. (E) Med12 does not affect Ring1b levels at the promoters of Med12+Ring1b target genes in mESCs. Ring1b is not enriched at promoters of Med12+Cdk8 target genes in mESCs. Chromatin immunoprecipitations (ChIPs) using chromatin from control (shNMC), Med12 (shMed12) and Ring1b (shRing1b) knockdown mESCs followed by qPCR on selected promoters. The enrichments are represented as percentage (%) of recovery over input and correspond to the average of 3 independent experiments +/− SEM ** P-values ≤ 0.007 as calculated by 2-tailed unpaired t test.
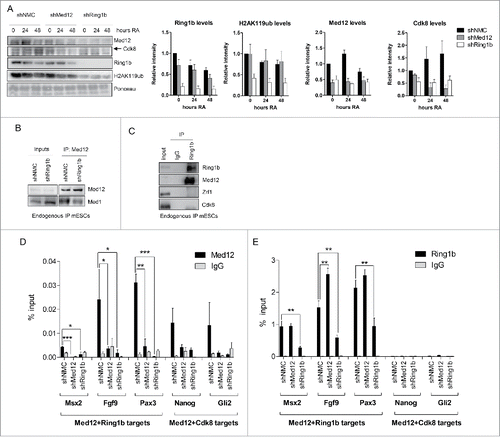
Figure 3. Ring1b and Med12 repress key developmental genes in mESCs. (A) Venn diagram of differentially expressed (DE) genes in control and knockdown mESCs. (B) GO analysis of biological functions of genes differentially expressed in both shMed12 and shRing1b mESCs. (C) Heatmap illustrating the expression profile of the genes differentially expressed in shMed12/shRing1b in control (shNMC), wild type, Cdk8 (shCdk8), Med12 (shMed12) and Ring1b (shRing1b) knockdown mESCs. (D) Knockdown of either Ring1b or Med12 causes de-repression of their common target genes in mESCs. Shown are the relative expression levels of selected Ring1b/Med12 target genes in the aforementioned cell lines. The relative mRNA levels are represented as fold change to the shNMC samples +/− SEM, n = 3. *** P-value< 0.0001, ** P-value < 0.001, * P-value < 0.02, as calculated by 2-tailed, unpaired t test. (E) Med12 and Ring1b directly control the expression of 116 genes. Venn diagram illustrating the overlap between differentially expressed genes in shMed12 and shRing1b mESCs and their common ChIP targets (). (F) Med12 and Ring1b repress the common target genes in mESCs. The plot depicts the expression Fold Change (FC) of the 116 direct target genes (). 100 out of 116 target genes (∼86%) are derepressed in both shRing1b and shMed12 mESCs.
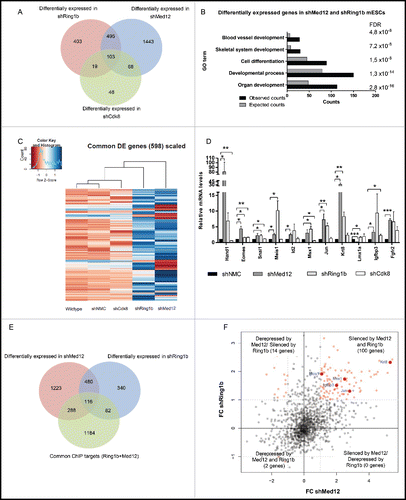
Figure 4. Med12 and Ring1b knockdown mESCs do not properly differentiate. (A) shMed12 mESCs exhibit low levels of pluripotency markers. Shown are the relative mRNA levels of pluripotency markers in shNMC, shCdk8, shMed12 and shRing1b mESCs. The relative mRNA levels are depicted as fold change to the shNMC samples +/− SEM, n = 3. *** P-value< 0.0001, ** P-value < 0.001, * P-value < 0.02, as calculated by 2-tailed, unpaired t test. (B) Embryoid Bodies (EBs) derived from shMed12 and shRing1b mESCs, have smaller diameters than those from shNMC and shCdk8 mESCs. Images of EBs were taken at 4x magnification at 2, 4, 6 and 8 d after the beginning of spontaneous differentiation by LIF removal. The size of the scale bar corresponds to 500 µm. (C-E) Knockdown of Med12 causes sustained repression of differentiation marker genes from the 3 germ layers. The mRNA levels of each marker gene are represented as fold change to the levels of the Day 0 sample of each genotype. Data are represented as a mean +/− SEM, n =3. *** P-value < 0.0001, ** P-value < 0.001, * P-value < 0.02, as calculated by 2-tailed, unpaired t test. (F) Developmental genes get upregulated upon RA-induced differentiation. The relative mRNA levels of Med12-Ring1b targets (Wnt8a-Sox7) and other RA-responsive genes (Hoxb4-Hoxd11) at 24hRA are represented as fold change to the untreated samples (mESCs) +/− SEM, n = 3. ** P-value = 0.009, *P-value < 0.05, as calculated by 2-tailed, unpaired t test. (G) RA-induced differentiation genes (significantly induced from 4F) are affected in the same (shMed12 = shRing1b) or opposite ways (shMed12 ≠ shRing1b) upon Med12 or Ring1b depletion in early differentiation (24hRA). The relative mRNA levels of the tested genes are represented as fold change to the shNMC samples +/− SEM, n = 3. *** P-value =0.0004, ** P-value ≤ 0.008, * P-value ≤ 0.04, as calculated by 2-tailed, unpaired t test.
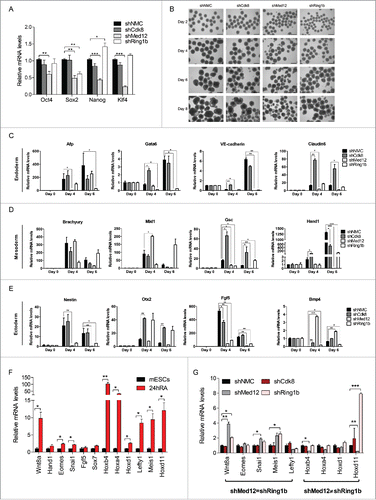
Figure 5. The Ring1b-Med12 target gene Hoxd11 is regulated by the ncRNA Dlx1as. (A) Med12 and Cbx7-PRC1 but not Cdk8, localize to Hoxd11 in mESCs. IGV Screenshot from RNAseq (mESCs) and ChIP-seq peaks illustrating the binding of Med12, Med1, Cdk8, Ring1b, Cbx7, RYBP and Suz12 at the Hoxd11 locus. (B) Illustration of the 2 Mb locus spanning the Dlx1as gene (green) and the HoxD cluster (red). The direction of transcription is indicated by the arrowheads. (C) The ncRNA Dlx1as regulates the expression of genes of the HoxD cluster in cis. mESCs were transfected with control siRNA or siRNA directed against Dlx1as and treated with RA. Twenty-four hours after RA administration the relative expression of genes in the 2 Mb locus were analyzed by qRT-PCR. Data are represented as a mean +/− SEM (n = 3). *** P-value<0.0001, ** P-value <0.01, * P-value < 0.02 as calculated by 2-tailed t test. (D) Med12 associates with Dlx1as in pluripotency and early differentiation. RIP experiments with Med12 antibodies and control IgGs from extracts of mESCs and differentiating cells (24h RA). The association of Dlx1as was analyzed in qRT-PCR experiments. Data are represented as a mean +/− SEM (n = 3). (E) Med12 associates with Dlx1as as a function of Ring1b. RIP experiments with Med12 antibodies and control IgGs using extracts from control and Ring1b knockdown mESCs. The association of Dlx1as, Hotairm (linc1548) and Rian was analyzed in qRT-PCR experiments. Data are represented as a mean +/− SEM (n = 3). The indicated P-value was calculated by F test to compare the differences. (F) Ring1b dissociates from Dlx1as upon differentiation. RIP experiments with Ring1b antibodies and control IgGs from extracts of mESCs and differentiating cells (24h RA). The association of the indicated ncRNAs Dlx1as, Hotairm and Rian was analyzed in qRT-PCR experiments. The figure represents an average of at least 3 independent experiments +/− SEM. The indicated P-value was calculated by F test to compare the differences. (G) The association of Ring1b and Med12 is RNA-mediated. Endogenous Ring1b IPs with nuclear extracts from wt mESCs. Precipitated material was subjected to RNaseA treatment prior to Western blotting and incubated with the indicated antibodies. Inputs represent 5% of the material used for the IPs. The relative intensity of Med12 Co-IP levels was calculated using the Ring1b levels as reference.
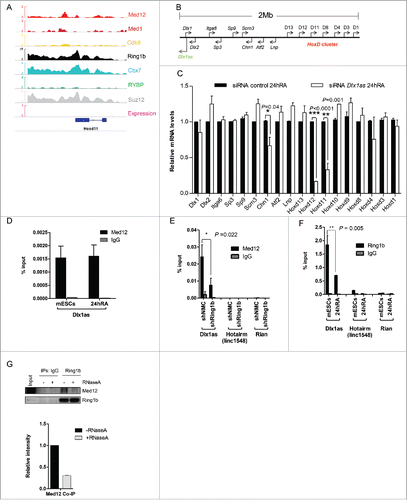
Figure 6. Zrf1 and Dlx1as activate Hoxd11 during differentiation. (A) Zrf1 is recruited to the promoter of Hoxd11 during early differentiation. ChIP experiments were carried out with FLAG antibodies and chromatin from stably transfected FLAG-Zrf1 mESCs either before or after RA administration (24h RA) and subsequent qRT-PCR. Data are represented as a mean +/− SEM (n = 3). (B) Knockdown of Zrf1 causes diminished activation of Hoxd11 in RA mediated mESC differentiation. 24 hours after RA administration the relative expression of genes in the 2Mb locus () was analyzed by qRT-PCR. Data are represented as a mean +/− SEM (n=3). *** P-value<0.0001, ** P-value <0.01, * P-value < 0.02 as calculated by 2-tailed t test. The depicted p-values were calculated by 2-tailed unpaired t test. (C) Zrf1 associates with Dlx1as during RA induced differentiation. RIP experiments with FLAG antibodies and control IgGs from extracts of stably transfected FLAG-Zrf1 mESCs before or after RA administration (24h RA). The association of the indicated ncRNAs, Dlx1as, Hotairm and Rian, was analyzed by qRT-PCR. Data are represented as a mean +/− SEM (n = 3). P-value = 0.004 as calculated by the F-test. (D) Schematic representation of Zrf1 showing the DnaJ, UBD and the c-terminal SANT domains. The mutated amino acids are indicated by asterisks. (E) Leucine 561 of the human ZRF1 is essential for RNA binding. Control, FLAG-tagged ZRF1 and FLAG-tagged mutant ZRF1 plasmids were expressed in HEK293T cells. After FLAG IP the precipitated material was analyzed by Westernblot and the co-precipitated RNA was analyzed with or without RNaseA treatment.
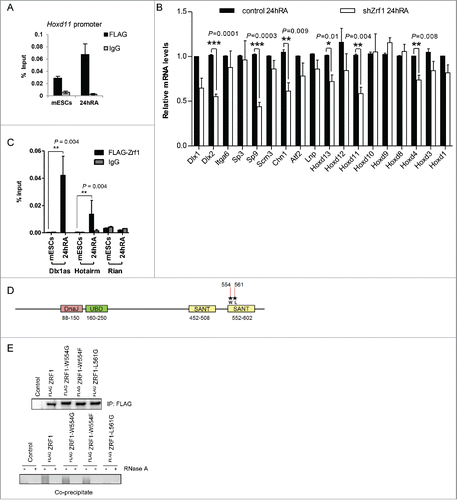
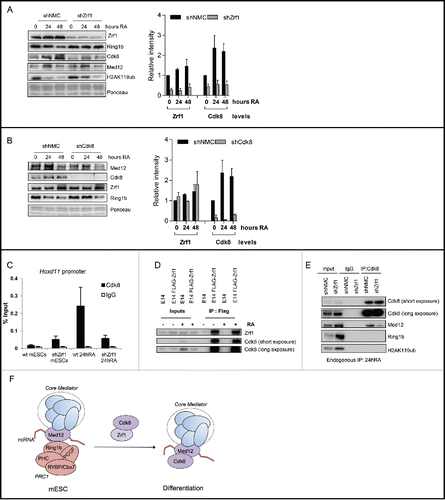
Figure 7. Zrf1 mediates the recruitment of Cdk8 to the Hoxd11 promoter. (A) Zrf1 displaces Ring1b from chromatin and facilitates Cdk8 recruitment. Chromatin from control or shZrf1 knockdown cells, either pluripotent or after RA administration was isolated and subjected to Western blotting and probed with the indicated antibodies. The relative intensities of Cdk8 and Zrf1 levels were quantified by using the ponceau intensity as reference and depicted as fold change to the 0 timepoint shNMC samples, +/− SEM, n = 3 (B) Knockdown of Cdk8 has no impact on Zrf1 recruitment to chromatin. Chromatin from control or shCdk8 knockdown cells, either pluripotent or after RA administration, was isolated and subjected to Western blotting and probed with the indicated antibodies. The relative intensities of Cdk8 and Zrf1 levels were quantified by using the ponceau intensity as reference and depicted as fold change to the 0 timepoint shNMC samples, +/− SEM, n = 3 . (C) Zrf1 controls the recruitment of Cdk8 to the Hoxd11 promoter during early differentiation. ChIPs were carried out with chromatin from nuclear extracts of wt and shZrf1 pluripotent and early differentiating mESCs utilizing Cdk8 antibodies or IgGs as control. Data are represented as a mean +/− SEM (n = 3). (D) Zrf1 interacts with Cdk8. Nuclear extracts from wildtype mESCs or mESCs stably expressing FLAG-Zrf1 were used in immunoprecipitations with FLAG antibodies. The precipitated material was subjected to Western blotting and incubated with the stated antibodies. (E) Zrf1 mediates the interaction of Cdk8 with Med12 during differentiation. Nuclear extracts from control or Zrf1 knockdown cells were used in endogenous IPs with Cdk8 antibodies. The precipitated material was subjected to Western blotting and incubated with the stated antibodies. (F) Proposed model for the transcriptional activation of Med12-Ring1b repressed genes. During pluripotency (mESCs) key developmental genes are repressed by PRC1 and Med12 probably also involving ncRNAs. During differentiation Zrf1 displaces PRC1 and recruits Cdk8 probably facilitating the assembly of the kinase module of Mediator.