Figures & data
Figure 1. Cell cycle analysis. Panel A – The graph shows the percentage of cycling (Ki67+) cells either in basal conditions or 6 and 48 hours post-irradiation.. Panel B -The picture shows a representative FACS analysis of irradiated (40 and 2000 mGy) and untreated MSCs. The assays were carried out 6 and 48 hours post-irradiation. Experiments were conducted in triplicate for each condition. The percentage of different cell populations (G1, S and G2/M) is indicated. Data are expressed with standard deviations. shR1, shR2 and sh107 cells were tested vs. control MSCs (shSCR, #p < 0.05). In each silenced condition (shSCR, shR1, shR2 and sh107), we compared irradiated versus unirradiated cells (*p < 0.05). The shSCR wild type MSCs; shR1, shR2 and sh107 are MSCs with silenced RB1, RB2/P130 and P107, respectively.
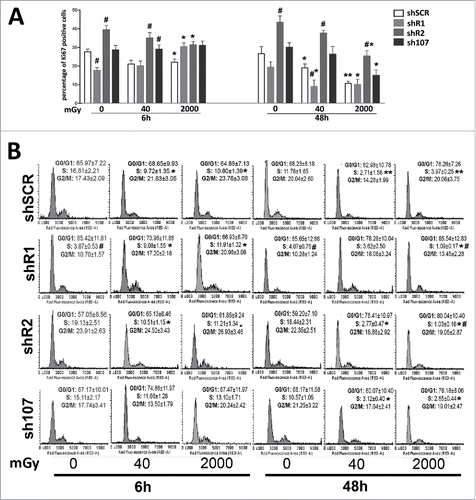
Figure 2. Gamma H2AX staining. Fluorescence photomicrographs show the merging of cells stained with anti-H2AX (green), anti Ki-67 (red) and DAPI (blue). Representative microscopic fields are shown. The graph shows the degree of H2AX phosphorylation. This was evaluated by counting the number of gamma-H2AX immunofluorescent foci per cell. Foci number was determined for 200 cells. Each dot represents an individual cell. Horizontal bars indicate mean value for each category. shR1, shR2 and sh107 cells were tested vs. control MSCs (shSCR, #p < 0.05). In each silenced condition (shSCR, shR1, shR2 and sh107), we compared irradiated versus unirradiated cells (*p < 0.05; **p < 0.01). The shSCR wild type MSCs; shR1, shR2 and sh107 are MSCs with silenced RB1, RB2/P130 and P107, respectively.
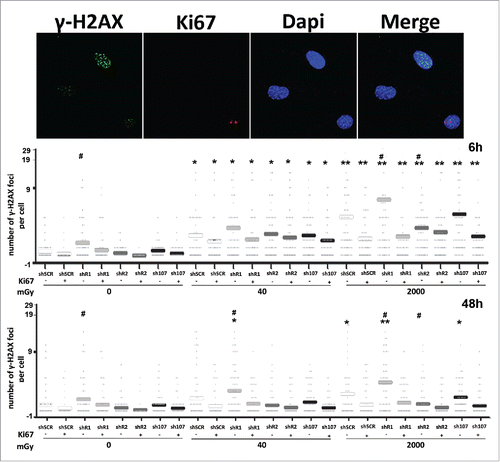
Figure 3. Analysis of DNA damage and repair. Fluorescence photomicrographs shows cells stained with anti-ATM (green) and Ki67 (red). Nuclei were counterstained with DAPI (blue). Representative microscopic fields are shown. For each silenced condition, the upper graph shows mean percentage of ATM(+) Ki67(−) resting cells either in basal conditions or 6 and 48 hours post-irradiation. The lower graph shows mean percentage of ATM(−) Ki67(+) cycling cells. Data are expressed with standard deviation. shR1, shR2 and sh107 cells were tested vs. control MSCs (shSCR, #p < 0.05). In each silenced condition (shSCR, shR1, shR2 and sh107), we compared irradiated versus unirradiated cells (*p < 0.05; **p < 0.01). The shSCR wild type MSCs; shR1, shR2 and sh107 are MSCs with silenced RB1, RB2/P130 and P107, respectively.
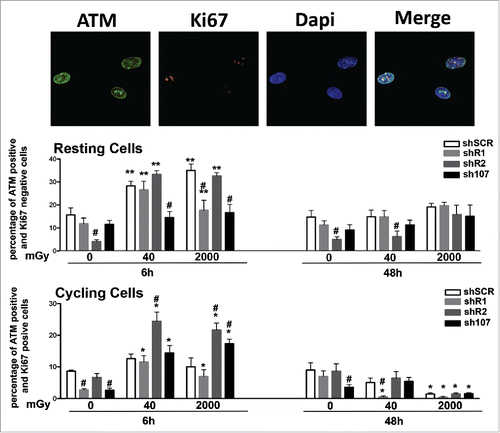
Figure 4. Evaluation of senescence and apoptosis. For each silenced condition, the upper graph shows the mean percentage of β-galactosidase positive cells either in basal conditions or 6 and 48 hours post-irradiation. The lower graph shows the mean percentage of Annexin V positive cells. Data are expressed with standard deviation. shR1, shR2 and sh107 cells were tested vs. control MSCs (shSCR, #p < 0.05). In each silenced condition (shSCR, shR1, shR2 and sh107), we compared irradiated versus unirradiated cells (*p < 0.05). The shSCR wild type MSCs; shR1, shR2 and sh107 are MSCs with silenced RB1, RB2/P130 and P107, respectively.
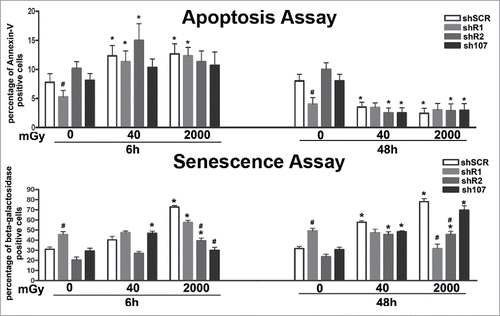
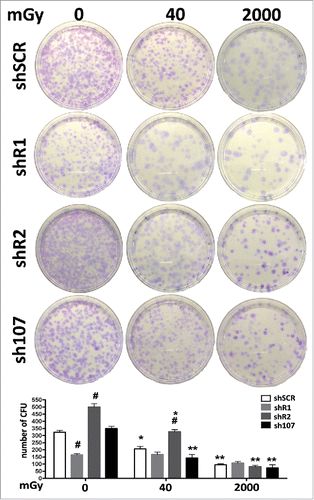
Figure 5. Stemness analysis by CFU assay. For each silenced condition, the pictures show representative crystal violet staining of clones obtained after 14 d of incubation. The graph shows the mean number of clones per 1,000 cells plated in a 100-mm dish. Data are expressed with standard deviations. shR1, shR2 and sh107 cells were tested vs. control MSCs (shSCR, #p < 0.05). In each silenced condition (shSCR, shR1, shR2 and sh107), we compared irradiated versus unirradiated cells (*p < 0.05; **p < 0.01). The shSCR wild type MSCs; shR1, shR2 and sh107 are MSCs with silenced RB1, RB2/P130 and P107, respectively.