Figures & data
Figure 1. Anti-21A RNAs transfection leads to a transient cell proliferation boost. (A) Quantification of red-labeled Anti-21A RNAs signal in S1 neuroblastoma cells at different time points (seven microscope fields were averaged for each time point). Microscope fields here reported represent S1 cells 24 hours after Anti-21A RNAs transfection. Scale bar: 200 μm. (B) Real time RT-PCR analysis of interferon response biomarkers in S1 cells transfected with Anti-21A RNAs or with water as control; gene expression levels are presented as normalized to GAPDH. Bulk Natural Killer population (NK) was used as positive control. (C) Population doubling of S1 cells transfected with Anti-21A RNAs or water as control, calculated in cell counting experiment. The inset represents the Population Doubling of S1cells replated 20 d after transfection with Anti-21A RNAs or water as control. (***p < 0.001).
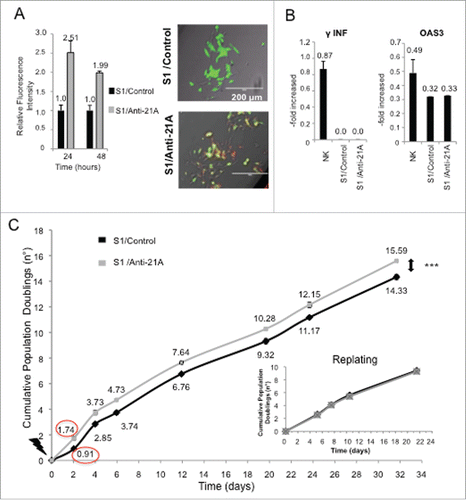
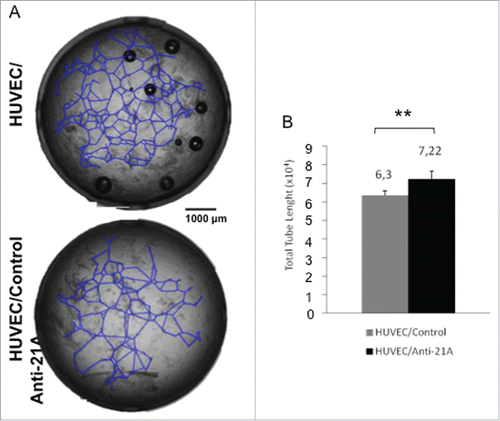
Figure 2. Anti-21A RNAs-dependent proliferation boost does not lead to the acquisition of tumorigenic potential. (A) Real time RT-PCR analysis of oncogenes and oncosuppressors in S1 cells transfected with Anti-21A RNAs; results (gray columns) are normalized to the water-transfected samples (black columns). (B) Colony-forming potential in Methocult of S1 cells transfected with Anti-21A RNAs and/or water (control). Scale bar: 1000 μm. (C) Real-time monitoring migration and invasion of S1 cells transfected with Anti-21A RNAs using the xCELLigence Migration and invasion assay. Blue, Huvec FBS- cells; green, S1 FBS-, Azure, S1 Anti-21A RNAs-transfected FBS-; magenta, HUVEC FBS+; violet, S1 FBS+; orange, S1 Anti-21A RNAs-transfected FBS+. Cells were seeded and their migration was monitored for 24 hours. Cells maintained in serum-free media served as a control. (D) Analysis of secreted cytokines in S1 cells transfected with Anti-21A RNAs (empty bars) or water control (full bars) by MILLIPLEX technology; the amount of cytokines secreted after a 2° treatment of Anti-21A are reported (striped bars). (E) Real time RT-PCR expression analysis of stemness markers; results are reported as Anti-21A/water control ratio. (*p < 0.05; **p < 0.01).
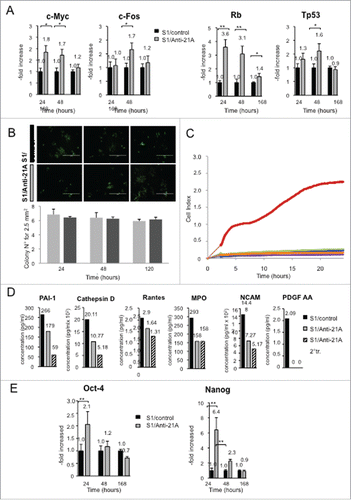
Figure 3. Anti-21A RNAs-transfected MSCs implanted in vivo drive the differentiation of an increased portion of compact fibrous tissue. A) [3H]-thymidine incorporation assay of hMSCs transfected with different combination of α, β and γ Anti-21A RNAs and B) effects on cell proliferation of different concentrations of the equimolar mix (α +β +γ) of Anti-21A RNAs. C) Reconstituted Hematoxilin/Eosin (H/E)-stained skelite sections of different implant conditions and their 10x respective magnifications. D) 10x magnification of Hematoxilin/Eosin-stained sections analyzed under transmitted light (T/L) and under polarized light (P/L). Scale bar: 250 μm. E) Picrosirius red/Fast green-stained skelite sections analyzed under transmitted light (T/L) microscopy and polarized light (P/L) microscopy. Mature collagen type I is stained in red, collagen type I in the maturation process in yellow, and collagen type III in light green in polarized light. All samples were obtained 45 d post-implantation. Scale bar: 250 μm.
![Figure 3. Anti-21A RNAs-transfected MSCs implanted in vivo drive the differentiation of an increased portion of compact fibrous tissue. A) [3H]-thymidine incorporation assay of hMSCs transfected with different combination of α, β and γ Anti-21A RNAs and B) effects on cell proliferation of different concentrations of the equimolar mix (α +β +γ) of Anti-21A RNAs. C) Reconstituted Hematoxilin/Eosin (H/E)-stained skelite sections of different implant conditions and their 10x respective magnifications. D) 10x magnification of Hematoxilin/Eosin-stained sections analyzed under transmitted light (T/L) and under polarized light (P/L). Scale bar: 250 μm. E) Picrosirius red/Fast green-stained skelite sections analyzed under transmitted light (T/L) microscopy and polarized light (P/L) microscopy. Mature collagen type I is stained in red, collagen type I in the maturation process in yellow, and collagen type III in light green in polarized light. All samples were obtained 45 d post-implantation. Scale bar: 250 μm.](/cms/asset/f5058d14-0063-4b6b-9cb5-f9c0cf912e3c/kccy_a_1181242_f0003_c.gif)
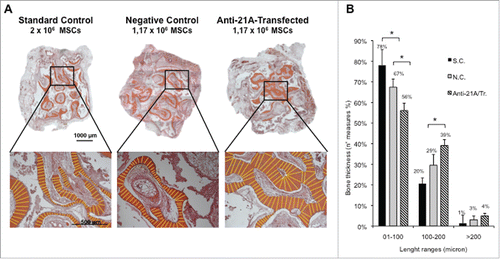
Figure 4. An increased bone mineralized area thickness is detected in implants from Anti-21A RNAs-transfected MSCs. (A) Reconstituted Hematoxilin/Eosin-stained skelite sections of different implant conditions and their respective magnifications. (B) Thickness distribution of mineralized tissue bone matrix deposition area. (SC: standard control group; NC: negative control group; Anti-21A/Tr: Anti-21A RNAs-transfected group).