Figures & data
Table 1. Saccharomyces cerevisiae strains used in this study.
Figure 1. Analysis of changes in the proteome of sit4Δ cells. Yeast extracts were prepared from S. cerevisiae BY4741 and sit4Δ cells grown in YPD medium to exponential phase. Proteins were separated by 2-dimensional gel electrophoresis and visualized by silver staining (A) or blotted into a nitrocellulose membrane. Immunodetection of proteins phosphorylated in serine residues was performed using an anti-phosphoserine antibody (B), as described in Materials and Methods. The experiment was reproduced 3 times, using independent samples. A representative gel/blot is shown. Arrows indicate proteins differentially expressed in sit4Δ cells compared to BY4741 cells.
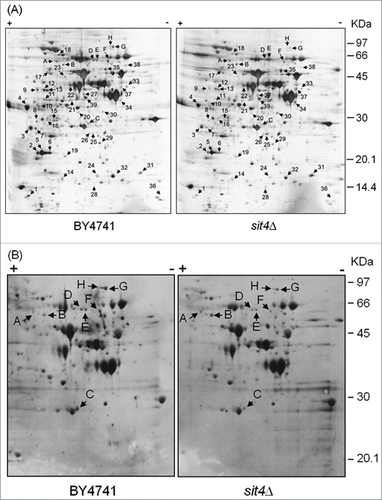
Table 2. Functional categories of proteins differentially expressed in sit4Δ cells. Proteins differentially expressed (sit4Δ vs parental cells) were sorted into functional categories according to MIPS (Munich information center for protein sequences). Proteins belonging to each category are indicated, together with their fold-change (in parentheses). Upregulated proteins are shown in bold.
Table 3. Identification of proteins differentially phosphorylated in sit4Δ mutants.
Figure 2. Hxk2p is hyperphosphorylated in cells with compromised CAPP or PP2A activity. Hxk2p-GFP was immunoprecipitated from the indicated mutants and immunodetected using anti-phosphoserine, anti-phosphotyrosine or anti-GFP antibodies, as described in Material and Methods. A representative experiment is shown.
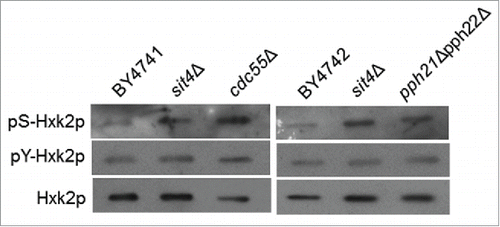
Figure 3. Hxk2p-S15 to Hxk2p-A15 mutation suppresses the phenotypes of sit4Δ cells. SIT4 gene was deleted in S. cerevisiae WAY.78-1 cells expressing wild type (Hxk2p-S15) or mutant (Hxk2p-S15A) hexokinase 2. (A) Oxygen consumption rates were measured as described in Materials and Methods using exponential phase cells grown in YPD medium. (B) Chronological lifespan was assessed by following the survival of cells maintained in the growth medium overtime. Cellular viability was measured at 2 to 3 d intervals and was expressed as % colony forming units (aged vs day 0). Values are mean ± SD of at least 3 independent experiments. (C) Oxidative stress resistance was assessed in cells were treated with 0.5 mM H2O2 for 30 min. Cell viability was determined by standard dilution plate counts and expressed as the percentage of the colony-forming units of non-stressed cells. Values are means ± SD of 3 independent experiments. *p < 0.05, **p < 0.01; Student's t-test. (D) Growth curves. (E) Cell cycle. Yeast cells were labeled with propidium iodide and analyzed by flow cytometry. The experiments were reproduced 3 times, using independent samples. A representative experiment is shown. (F) Quantification of the percentage of cells in the different phases of the cell cycle [from (E)].
![Figure 3. Hxk2p-S15 to Hxk2p-A15 mutation suppresses the phenotypes of sit4Δ cells. SIT4 gene was deleted in S. cerevisiae WAY.78-1 cells expressing wild type (Hxk2p-S15) or mutant (Hxk2p-S15A) hexokinase 2. (A) Oxygen consumption rates were measured as described in Materials and Methods using exponential phase cells grown in YPD medium. (B) Chronological lifespan was assessed by following the survival of cells maintained in the growth medium overtime. Cellular viability was measured at 2 to 3 d intervals and was expressed as % colony forming units (aged vs day 0). Values are mean ± SD of at least 3 independent experiments. (C) Oxidative stress resistance was assessed in cells were treated with 0.5 mM H2O2 for 30 min. Cell viability was determined by standard dilution plate counts and expressed as the percentage of the colony-forming units of non-stressed cells. Values are means ± SD of 3 independent experiments. *p < 0.05, **p < 0.01; Student's t-test. (D) Growth curves. (E) Cell cycle. Yeast cells were labeled with propidium iodide and analyzed by flow cytometry. The experiments were reproduced 3 times, using independent samples. A representative experiment is shown. (F) Quantification of the percentage of cells in the different phases of the cell cycle [from (E)].](/cms/asset/85310855-0d83-4c4e-a31f-95383cfd8673/kccy_a_1183846_f0003_b.gif)
Figure 4. Hxk2p is hypophosphorylated in isc1Δ cells by a Sit4p-dependent mechanism. (A) Yeast extracts were prepared from S. cerevisiae BY4741, isc1Δ and isc1Δsit4Δ cells and a phosphoproteome analysis was performed, as described in legend to . The region of the blots showing Hxk2p and Dug1p was selected. Spot intensities were quantified by densitometry. Values are means ± SD of 3 independent experiments. *p < 0.05. (B) Chronological lifespan of S. cerevisiae hxk2Δ and isc1Δhxk2Δ cells expressing Hxk2p-S15 or Hxk2p-S15E. Cellular viability was determined as described in legend to . Values are means ± SD of 4 independent experiments.
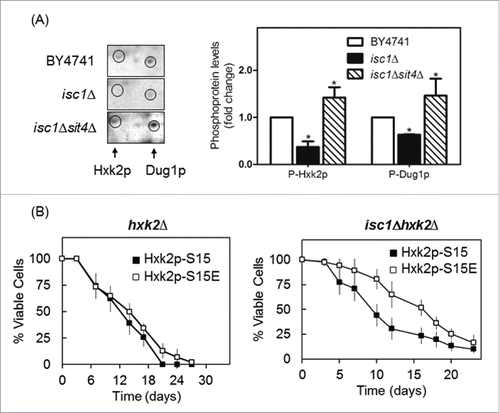
Figure 5. Snf1p activation does not mediate Hxk2p phosphorylation in sit4Δ cells. (A) Protein extracts from S. cerevisiae BY4741, sit4Δ and snf1Δ cells were analyzed by immunoblotting using anti-Snf1 or anti-AMPK (phospho T172) as described in Materials and Methods. (B) Hxk2p-GFP was immunoprecipitated from protein extracts of S. cerevisiae BY4741, sit4Δ, snf1Δ and sit4Δsnf1Δ cells and analyzed by immunoblotting using anti-GFP or anti-phosphoserine as described in Materials and Methods. Representative blots are shown (out of 3 independent experiments).
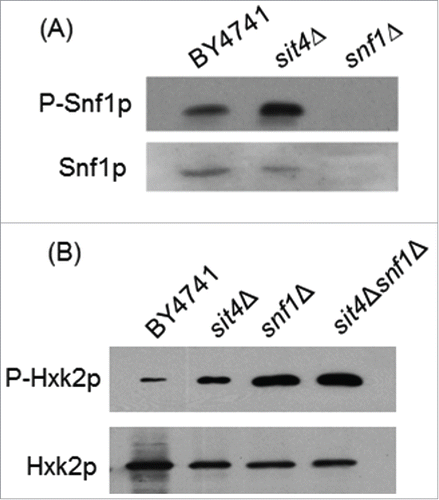
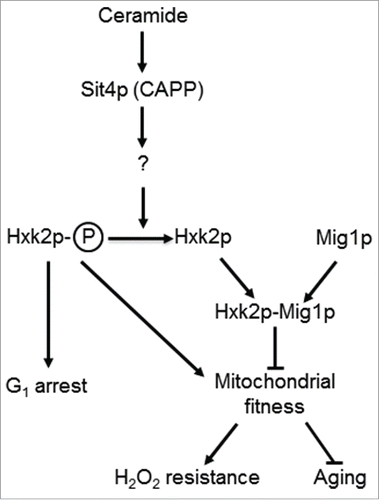
Figure 6. A model for the role of Sit4p in the regulation mitochondrial function. In parental cells, dihydroceramides (dh-Cer) and phytoceramide (phyto-Cer) levels are kept low, preventing the dephosphorylation of hexokinase 2 (Hxk2p) in serine-15, indirectly regulated by the ceramide-activated protein phosphatase Sit4p. In sit4Δ cells, the increase of Hxk2p phosphorylation contributes to mitochondrial fitness, increasing oxidative stress resistance and chronological lifespan. In isc1Δ cells, the higher levels of dh-Cer- and phyto-Cer activate Sit4p leading to Hxk2p dephosphorylation. This results in mitochondrial dysfunction, leading to a disturbed redox homeostasis, premature aging and oxidative stress sensitivity. Other proteins regulated by Sit4p may also contribute to these phenotypes.