Figures & data
Figure 1. Mekk1 regulates Cdkn1b expression in Th17 cells. (A) Naïve CD4+T cells were extracted from WT and Map3k1ΔKD mice and differentiated under Th17 conditions. FACS analysis was performed with the indicated antibodies (4 mice per experiment). (B) Quantitation of WT and Map3k1ΔKD IL-17+CD4+T cells (▪ WT and ▪ Map3k1ΔKD) differentiated under Th17 conditions (n = 3). (C) Naïve CD4+ T cells were extracted from WT and Map3k1ΔKD mice and incubated with SP600125, NSC697923 or SB431542 inhibitors while differentiating under Th17 conditions (4 mice per experiment). FACS analysis was performed with the indicated antibodies. (D) Global gene expression analysis of Th17 cells. Naïve CD4+ T cells were extracted from WT and Map3k1ΔKD mice and differentiated into Th17 cells. RNA was isolated from WT and Map3k1ΔKD splenic Th17 cells, processed and hybridized onto Affymetrix arrays. Bioinformatics analysis was performed and a heat map comparing gene hits between WT and Map3k1ΔKD Th17 cell microarray screen hits was constructed. The data is from 3 independent experiments (4 mice per experiment). (E) Real-time PCR analysis of Map3k1ΔKD for Cdkn1b undergoing Th17 differentiation. Naïve CD4+ T cells from the spleen of WT and Map3k1ΔKD mice were isolated, differentiated under Th17 conditions and their RNA was analyzed by real-time PCR as indicated (▪ WT and ▪ Map3k1ΔKD). The average relative expression (± SEM) of genes from 3 independent experiments was statistically analyzed, where appropriate, by 2-tailed Student's t test (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001).
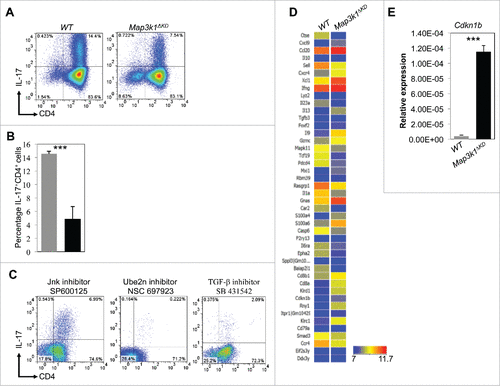
Figure 2. Mekk1 and Tak1 regulate iNKT cell proliferative expansion in the liver. (A) Liver cells from WT, Map3k1ΔKD, LckCre/+Map3k1f/f and LckCre/+Map3k7f/f mice were isolated and stained with CD1d tetramer and anti-CD3 antibody and analyzed by flow cytometry as shown. Data is representative of at least 5 mice. (B) Aberrant iNKT liver cell expansion in LckCre/+Map3k1f/fand LckCre/+Map3k7f/f(![]()
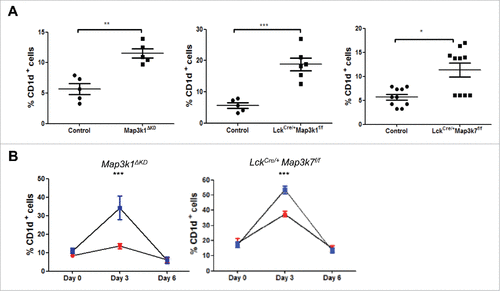
Figure 3. Schematic illustrating Mekk1 signaling and the regulation of Cdkn1b expression in T cells. Following antigen receptor engagement Mekk1 signaling activates the Jnk Mapk pathway to regulate Cdkn1b transcription. p27KipCitation1 negatively regulates T cell proliferative expansion.