Figures & data
Figure 1. Analysis of the HMGI-(C)mRNA expression in breast cancer patients. HMGI-C mRNA expression levels evaluated in 12 marginal tissues, 12 tumoral tissues, MDA-MB-468 and HFF-1 cells. ****p < 0.0001 vs. marginal tissues group and HFF-1 cells.
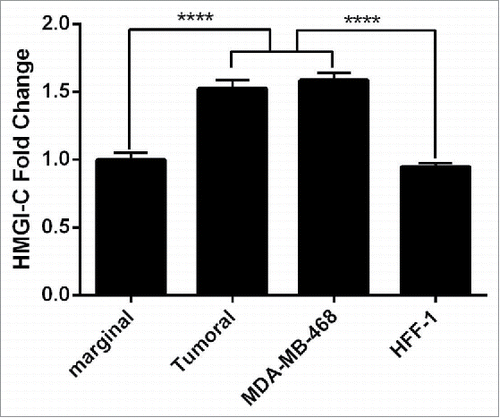
Figure 2. Suppression of HMGI-(C)mRNA and protein expression by siRNA in breast adenocarcinoma. HMGI-C mRNA expression in MDA-MB-468 cells, which were transfected with specific siRNA or (NC) siRNA after (A) 24, 48, 72 h, and (B) 40 pmol, 60 pmol, 80 pmol of specific siRNA. Relative HMGI-C mRNA expression was measured by qRT-PCR using 2(-ΔΔCt) method. (C) A representative protein gel blot of b-actin and HMGI-C proteins from cells transfected with HMGI-C siRNA or (NC) siRNA. (D) The expression level of each band was quantified using densitometry and normalized to the respective B-actin. The results were expressed as mean ± SD (n = 3); *p < 0.05, **p < 0.001, *** p = 0.0001, ****p < 0.0001 versus control group.
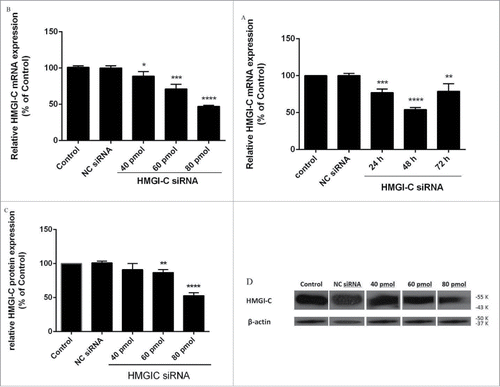
Figure 3. Relative Let-7a (A) and miR-34a (B) expression levels were measured by qRT-PCR using 2(-ΔΔCt) method. **p < 0.001, ****p < 0.0001 vs. control group.
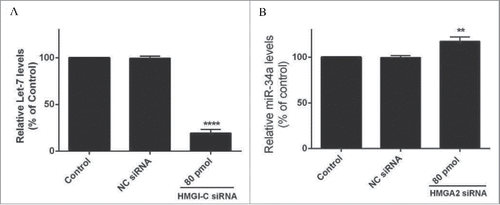
Figure 4. Effect of HMGI-C siRNA on the MDA-MB-468 cells. 48 h after transfection with HMGI-C siRNA (40, 60, 80 pmol), cytotoxicity of treatments was determined by MTT assay. The results were expressed as mean ± SD (n = 3); **P < 0.01, ***P < 0.001 versus control group.
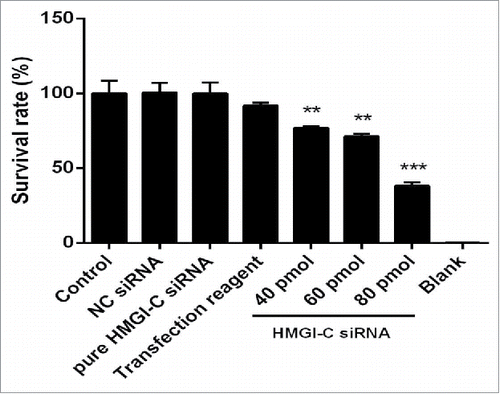
Figure 5. siRNA-mediated targeting of HMGI-C strongly sensitized MDA-MB-468 cells by stimulation of apoptosis. Untreated (A), and 80 pmol HMGI-C treated MDA-MB-468 (B) were prepared and assayed with TUNEL assay. FACS analysis of cell distribution for apoptosis and necrosis on MDA-MB-468 cells for untreated (C), (NC) siRNA (D) and 80 pmol siRNA (E), prepared after 48 h. Percentage of dual-positive (Annexin V and PI positive) cells from 3 independent experiments were quantified and presented as mean ± SD (n = 3). (F) Relative P53, caspase3, 9, 8, Bcl2 (5G, 5H) protein expression were evaluated by immune-blotting and densitometry using imageJ software. *p < 0.05, ****p < 0.0001, vs. control group.
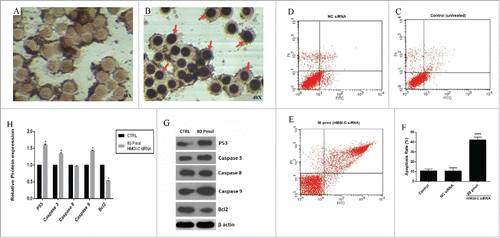
Figure 6. siRNA-mediated targeting of HMGI-C, prolonged G0/G1 arrest in MDA-MB-468 cells. Untreated, (NC) siRNA and 80 pmol HMGI-C treated cells were prepared and stained with the propidium iodide (PI). The percentage of the cell population in untreated, negative-control siRNA and 80 pmol HMGI-C siRNA group evaluated in each phases. The results are expressed as mean ± SD (n = 3); *p < 0.05, versus control group.
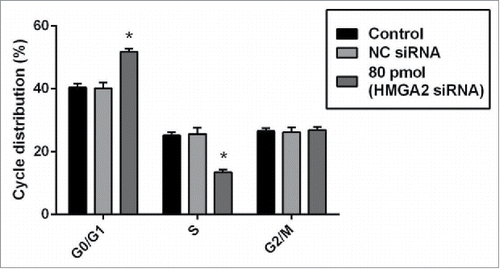
Table 1. Primer sequences.