Figures & data
Figure 1. Effect of RNAi mediated PGRMC1 silencing on bGC growth. (A) Graph showing PGRMC1 mRNA silencing in PGRMC1 RNAi treated bGC as assessed by qRT-PCR. PGRMC1 expression level was normalized using GAPDH as reference gene. Data were analyzed by one way ANOVA, followed by Tukey's Multiple Comparison Test, Values are means ± SEM (n = 3). *indicates significant differences between groups (P < 0.05). (B) Representative Western blot showing the decreased expression of all PGRMC1 bands at 72 h in PGRMC1 RNAi treated bGC; β tubulin was used as loading control. (C) Graph showing the effect of PGRMC1 down regulation on cell growth. Data were analyzed by two way ANOVA followed by Bonferroni post-hoc test. Values are means ± SEM (n = 3). *indicates significant differences between groups P < 0.05. (D) Graph showing the increase in the percentage of cells arrested at G2/M phase after 72 h from transfection, as assessed by flow cytometry analysis. Data were analyzed by unpaired Student's t-test, Values are means ± SEM (n = 3). *indicates significant differences between groups (P < 0.05).
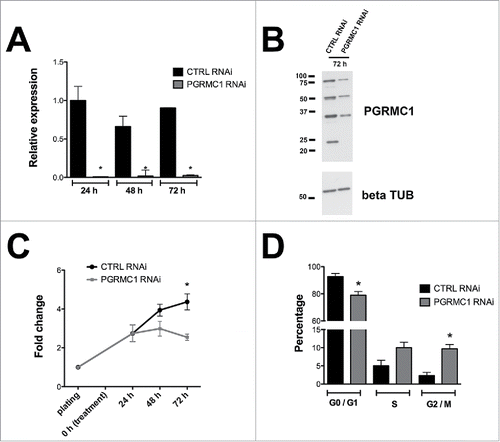
Figure 2. Effect of PGRMC1 silencing on bGC mitosis by time-lapse analysis of transfected bGC. (A) Example of normal mitosis occurring in bGC transfected with CTRL RNAi. The cell goes through all the mitotic phases from prophase to telophase giving rise to two daughter nuclei (see Supplemental Movie 1). (B) Example of abnormal mitosis in bGC transfected with PGRMC1 RNAi, in which the cell starts the division process undergoing prophase but then fails to proceed beyond the Ana/Telophase (see Supplemental Movie 2). Additional examples of abnormal mitosis occurring in PGRMC1 RNAi treated bGC are shown in Supplemental Movies 3 and 4. (C) Graph showing the frequency of normal and aberrant mitotic events assessed in CTRL and PGRMC1 RNAi treated cells. Data were analyzed by Fisher exact test. *indicates significant differences between groups P < 0.05. (D) Graph showing the time to complete mitosis in CTRL and PGRMC1 RNAi treated cells. Data were analyzed by unpaired Student's t-test, *indicates significant differences between groups (P < 0.05). This experiment was replicated four times; the total number of cells analyzed is shown in brackets.
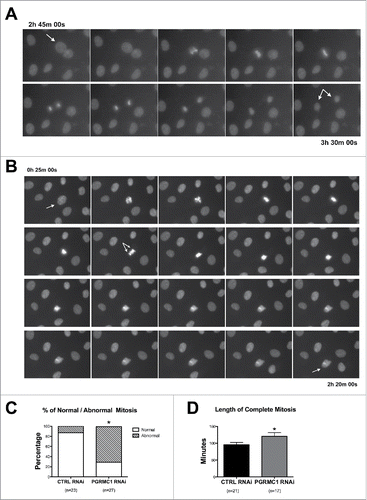
Figure 3. Colocalization PGRMC1-AURK in bGC. Representative images showing PGRMC1 (red) and AURKB (green) colocalization during the different mitotic phases of cultured bGC; DNA was stained with DAPI (blue). PGRMC1 and AURKB start to colocalize to the mitotic spindle region during prophase and the colocalization is more pronounced in the central spindle during ana/telophase. Scale bar is 10 µm.
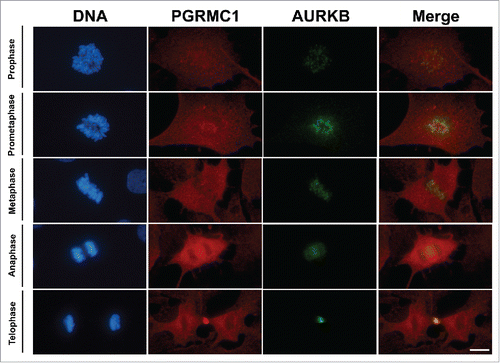
Figure 4. Interaction between PGRMC1 and AURKB during mitosis in bGC. (A) Representative images of bGC showing PGRMC1 - AURKB interaction as assessed by In situ proximity ligation assay (PLA) from prophase until telophase. DNA was stained with DAPI (white). The red spots indicate PGRMC1-AURKB interactions. Scale bar is 10 µm. (B) Graph showing the increased interaction between these two proteins during telophase. Data were analyzed by one way ANOVA followed by Tukey's Multiple Comparison Test. Values are means ± SEM; *indicates significant differences between groups (P < 0.05). This experiment was replicated three times with the total number of cells analyzed shown in brackets.
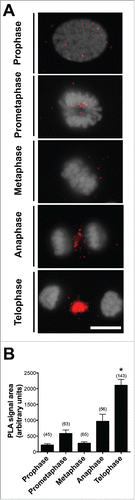
Figure 5. Effect of AG205 treatment on PBI emission. (A) Representative images showing in vitro matured oocytes with (left, arrow) or without (right) extruded PBI. (B, C) Graphics showing the effect of AG205 treatment on the percentage of oocytes that extruded the PBI in COC and DO, respectively. Data were analyzed by one way ANOVA followed by Tukey's Multiple Comparison Test. Values are means ± SEM (n = 3); a,b different letters indicate significant differences between groups (P < 0.05).
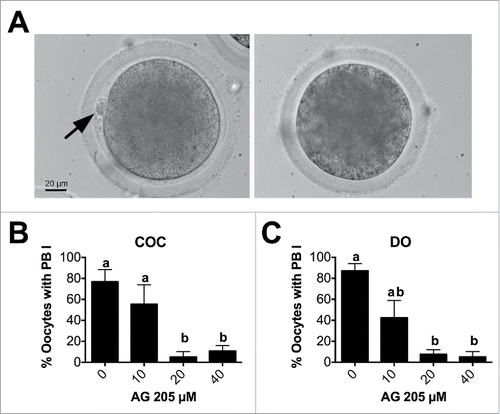
Figure 6. Effect of AG205 treatment on meiotic progression. (A) Representative images showing oocytes classified as MI, Ana/Telophase, MII, aberrant or degenerated after AG205 treatment. Aberrant mitotic figure shown in this figure is characterized by the presence of DNA scattered within the ooplasm (arrow), while additional examples of aberrant meiotic figures observed after AG205 treatment are shown in Figure S1. DNA was stained with DAPI (white). Scale bar is 50 µm. Insets show the DNA at 2X magnification. Graphs in (B) and (C) show the percentages of oocytes at each stage of the first meiotic division (shown in A) in COC and DO respectively. Data were analyzed by one way ANOVA followed by Tukey's Multiple Comparison Test. Values are means ± SEM (N = 3); a,b different letters indicate significant differences between groups (P < 0.05).
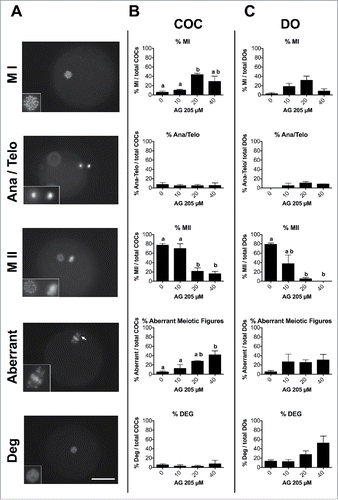
Figure 7. Effect of AG205 on PGRMC1 and AURKB localization. Representative images showing PGRMC1 and AURKB localization in MII plates of matured oocytes or in oocytes showing aberrant meiotic figures. COC were treated with 0 or 20 µM AG205 for 24 h. After AG205 treatment oocytes were fixed, immunostained with anti- PGRMC1 (red) and AURKB (green) antibodies; DNA was stained with DAPI (blue). A total of 97 oocytes from 2 independent experiments were analyzed. Both PGRMC1 and AURKB showed a focused centromeric localization in oocytes with MII plate, while they often showed a more diffused localization in aberrant meiotic figures with scattered chromosomes. When clumps of chromatin were present within the ooplasm, none of them were associated with AURKB and/or PGRMC1. Finally, when double meiotic plates were present, both AURKB and PGRMC1 showed a focused localization on metaphasic chromosomes of both plates.
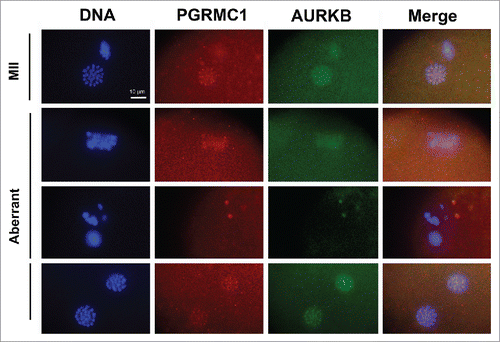
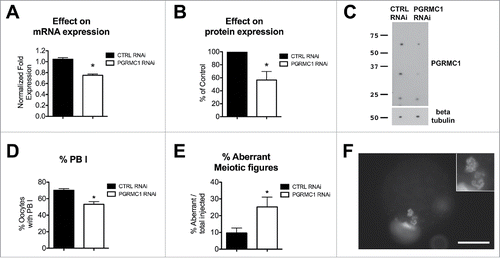
Figure 8. Effect of PGRMC1 RNAi treatment on PGRMC1 expression level, PBI extrusion and meiotic progression. (A) Graph showing PGRMC1 mRNA expression level in CTRL and PGRMC1 RNAi treated oocytes by RT-qPCR; PGRMC1 expression level was normalized using GAPDH and HIST1H2A as reference genes and differences in gene expression levels were assessed with the Delta-Delta Ct method. Data were analyzed by Student's t-test. Values are means ± SEM (N = 3); * indicates significant differences between groups (P < 0.05). (B and C) graph and representative Western blotting showing PGRMC1 protein expression levels in CTRL and PGRMC1 RNAi treated oocytes; β tubulin was used as loading control. PGRMC1 protein expression in PGRMC1 RNAi treated oocytes is expressed as a percentage of the CTRL RNAi treated group. Data were analyzed by one sample t-test. Values are means ± SEM (n = 6). *indicates significant differences (P < 0.05). (D) Graph showing the percentage of oocytes that extruded the PBI after CTRL or PGRMC1 RNAi treatment and IVM. Data were analyzed by t-test. Values are means ± SEM (n = 14); *indicates significant difference between groups (P < 0.05). (E) Graph showing the percentage of oocytes showing aberrant meiotic figures. Data were analyzed by t-test. Values are means ± SEM (n = 6); *indicate significant differences between groups (P < 0.05). Image in (F) is representative of an aberrant meiotic figure observed after PGRMC1 RNAi treatment, in which chromosomes and DNA clumps are dispersed in the ooplasm and not organized in a MII plate (additional examples of aberrant meiotic figures are shown in Fig. S3). Scale bar is 50 µm. Insets show the DNA at 2X magnification.