Figures & data
Figure 1. Flowchart depicting the strategy to identify core GECN in HeLa and ES cells. In this study, we mined omics information from big databases, such as transcription and miRNA regulation interactions, to construct real GECNs using genome-wide high-throughput data, a dynamic model, and AIC. For drug screening, information was integrated from cervical carcinogenic mechanisms in cancer-specific GECN and drug databases for cervical cancer treatment.
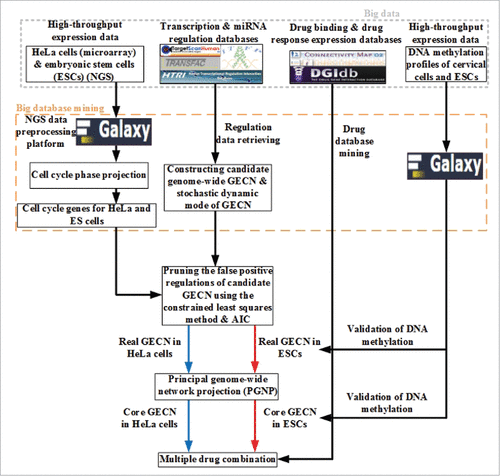
Figure 2. Identification of HeLa and ESC cell cycle genes after applying the cell cycle projection method. HeLa and ES cells cell cycle genes were selected according to the maximal phase-specific ability value, i.e., ai,k* in (3). The right bar represents the Z score, from maximum (white) to minimum value (black). The phase-shift of cell cycle genes can be observed as white blocks along the diagonal.
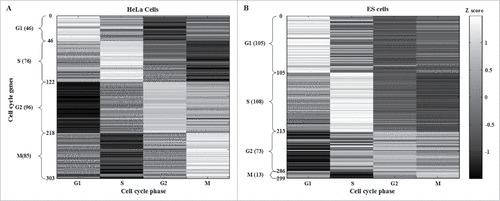
Table 1. Top 5 phase-specific genes in HeLa and ES cells.A complete list of identified cell cycle genes in HeLa cells (303) and ESCs (299) can be found in Tables S1 and S2, respectively.
Figure 3. Real GECNs of HeLa and ES cells. The number of G1 and S phase TFs and genes, and G2 phase TFs decreased significantly in HeLa compared to that in ES cells; whereas the number of M phase TFs and genes increased significantly. In addition, the numbers of G1, S, and G2 phase TF regulations in HeLa cells also decreased compared to ES cells; whereas the number of M phase TF regulations increased. These results support the immortal nature of HeLa cells being due to dysregulation of genes at the G1/G2 checkpoint and cell cycle exit.
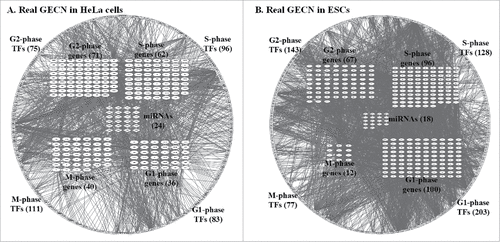
Figure 4. Specific and common core GECNs of HeLa cells and ESCs. Specific and common core networks were selected by PGNP with 0.001 (HeLa cells) and 0.1 (ESCs) thresholds. For each core network, inner circles contain miRNAs; middle circles correspond to cell cycle genes; and outer circles include TFs.
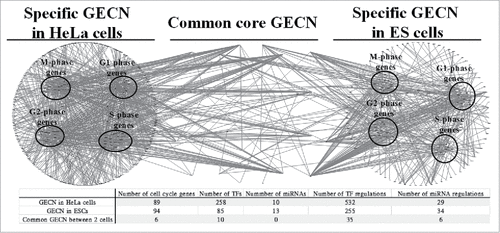
Figure 5. Principal networks of HeLa (A) and ES (B) cells. To find the principal networks, we assumed that they contained cell cycle genes with top PGNP projection values and that each of the 4 cell cycle phase groups involved at least one cell cycle gene. We identified the principal GECNs from the specific GECNs shown in . Genes/TFs/miRNAs in blue and red denote activated expression in HeLa and ES cells, respectively (p-value ≤ 0.05).
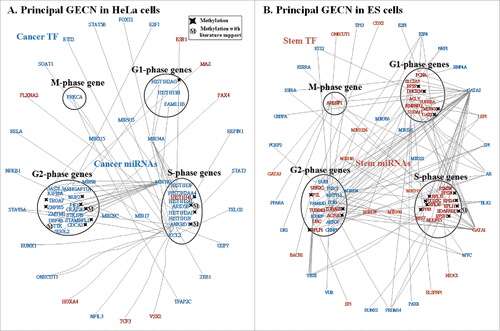
Figure 6. Carcinogenic and stemness mechanisms. (A) Carcinogenic mechanism of HeLa cells and (B) stemness mechanism of ES cells. Carcinogenic and stemness mechanisms were derived by applying big mechanism analysis to the principal GECN in HeLa and ES cells, respectively (), using the gene ontology tool DAVID. Genes/TFs/miRNAs in blue and red denote activated expression in HeLa and ES cells, respectively (p-value ≤ 0.05).
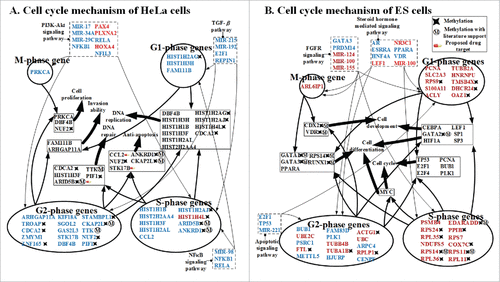
Table 2. Proposed multiple drugs for the treatment of cervical cancer.Proposed targets and structures of methotrexate, quercetin, and mimosine derived from the ZINC database. The identified drugs suppress activated cell cycle genes, such as ARID5B, STK17B, and CCL2, without affecting 6 common core genes, PLK1, TOP2A, AURKB, FAM83D, HJURP, and HMMR. The ranked list of all drugs in CMap is shown in Table S5. Methotrexate targets abnormal methylation in S phase, quercetin inhibits anti-apoptosis in G2 phase, and mimosine blocks most signal transduction and inhibits anti-apoptosis in S phase. Apoptosis reduces accumulated genetic mutations, abnormal DNA methylations, and the dysregulation of miRNAs in normal cells; thus, achieving carcinogenesis inhibition.
