Figures & data
Figure 1. Expression of MAD2 isoforms in multiple cancer cell lines and primary non-cancerous foreskin fibroblasts. The three MAD2 isoforms were simultaneously amplified by pairing forward and reverse oligonucleotides to exons 1 and 5, respectively (). β-actin was used as an internal control. The three isoforms were differentially expressed in different cancer cell lines and in non-cancerous primary foreskin fibroblasts (Fib-P4 and Fib-P9).
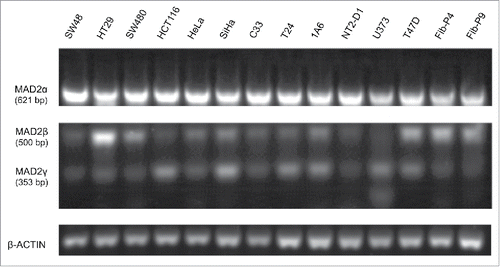
Figure 2. The sequences of the 3 MAD2 isoforms. (A) The coding sequence for MAD2L1 contains a 618-bp open reading frame (pale pink bar) consisting of 5 exonsCitation22 that are translated into a 205-amino acid protein. (B) MAD2β, first described by Yin et al.,Citation20 does not contain exon 3 of MAD2. The transcript contains a 273-bp open reading frame that is translated into a 90-amino acid protein. (C) In MAD2γ, exons 2 and 3 are also removed by alternative splicing. MAD2γ contains a 126-bp open reading frame, which is predicted to encode a 41-amino acid peptide.
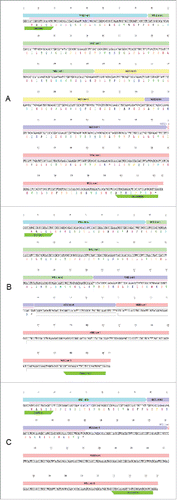
Figure 3. Ectopic expression of MAD2γ in the SAC-competent colorectal cancer cell line HCT116. (A) MAD2γ mRNA expression was determined by semiquantitative RT-PCR using specific primers pairing exon 1 (forward) and the spliced region between exon 1 and exon 4 (reverse). (B) Protein levels were detected by Western blotting using the MAD2 polyclonal antibody N-19, which recognizes the N-terminus of MAD2. (C) When cells were treated with 100 nM Taxol for 12 h, MAD2γ overexpression significantly decreased the mitotic index compared to non-transfected cells (HCT116-Tx, P = 0.03) and cells transfected with empty vector (HCT116/pcDNA-Tx, P = 0.0001).
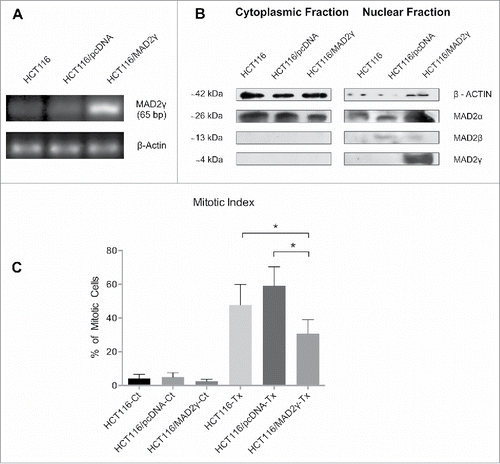
Figure 4. In silico identification of a MAD2-interacting motif present in the C-terminal domains of MAD2 isoforms. (A) Ribbon diagram of the crystal structure of the active conformation of MAD2α (C-MAD2α) (PDB ID 1GO4) and a homology model of the isoforms MAD2β and MAD2γ. (B) Exon arrangements (above) and secondary structures of the MAD2α and MAD2 isoforms (below). Alternative splicing produces 2 MAD2 isoforms: MAD2β and MAD2γ, from which exon 3 and exon 2 and 3 are excluded, respectively. A frameshift occurs in exon 4 causing a premature stop codon and the creation of a distinct C-terminus peptide (shown in dark blue). (C) Sequence alignment of the C-terminal peptides of MAD2 isoforms with the MAD2-interacting motifs (MIMs) of CDC20 proteins from various organisms (Hs, Homo sapiens; Mm, Mus musculus; Bt, Bos taurus; Xl, Xenopus laevis). (D) Schematic drawing of SAC inhibition by MAD2 isoforms. Upon SAC activation, MAD1/C-MAD2α catalyzes the conversion of the cytosolic conformation of MAD2α (O-MAD2α) to C-MAD2α. We propose that MAD2 isoforms could compete with CDC20 for C-MAD2α binding. Because the MAD2(β/γ)/C-MAD2 complex cannot bind to CDC20, free CDC20 can activate APC/C and induce the onset of anaphase.
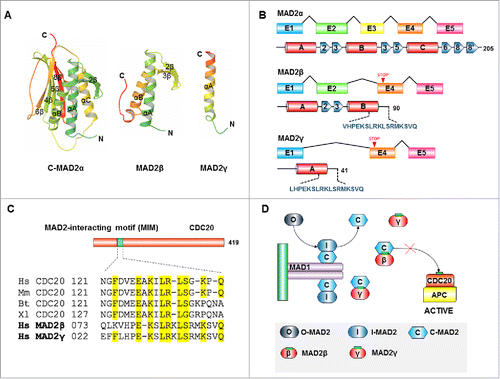
Figure 5. Subcellular localization of MAD2γ. HCT116 cells were transiently transfected with the Flag-MAD2γ construct. After 48 h, the cells were fixed, and MAD2γ was immunostained with a FITC-coupled-anti-DDDK tag (ab1259, Abcam, Biotech Co., Cambridge, UK). MAD2γ localized to the nucleus.
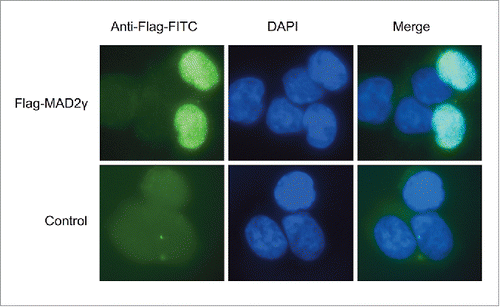
Figure 6. MAD2γ and MAD2α expression in patients with TGCTs. (A) ROC curve analysis and the Youden index were used to determine the optimal cut-off point for distinguishing overexpression and underexpression of MAD2γ and MAD2α between cisplatin-sensitive and cisplatin-resistant patients. The cut-off value for MAD2γ was 3.091, and for MAD2α it was 1.33. (B) and (C) Relative quantification of MAD2γ and MAD2α expression levels between both groups of patients as measured by quantitative real-time RT-PCR. MAD2γ was overexpressed in 70% of the cisplatin-resistant patients, whereas only 30% of the cisplatin-sensitive patients overexpressed MAD2γ. There was no significant difference in MAD2α expression between the groups of patients.
Table 1. Clinical and histopathological characteristics of patients with TGCT
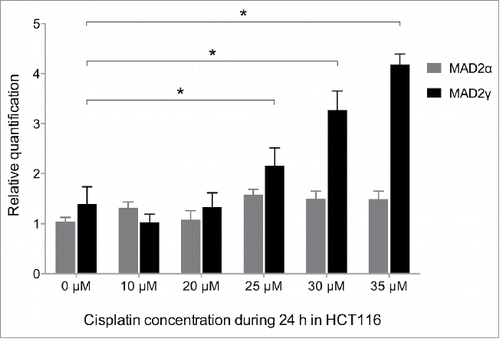
Figure 7. MAD2γ and MAD2α expression levels change in response to treatment with cisplatin. The colorectal cancer cell line HCT116 was exposed to different concentrations of cisplatin for 24 h. MAD2γ and MAD2α expression levels were then measured by quantitative real-time RT-PCR. The cells exhibited a significant increase in MAD2γ expression, especially when treated with concentrations close to the IC50 value (IC50 = 29 µM). In contrast, the expression of MAD2α did not significantly change.