Figures & data
Figure 1. Blocking antibody targeting integrin αv blocks re-epithelialization in an organotypic wound healing model. (A) Images of 3D printed runway for organotypic re-epithelialization assay. (B) Timecourse for organotypic re-epithelialization assay. Keratinocytes are seeded onto the left side of the runway on day 0, and the right side of the runway is blocked until day 5. On day 5, blocking antibodies are added, and keratinocytes migrate over the course of 5 days, until the tissue is harvested on day 10. (C) Visualization of re-epithelialization assay upon treatment with control (Ms IgG) antibody or L230 antibody. (D) Representative Hematoxylin & Eosin (H&E) stain for tissue shown in C, harvested at day 10. Arrow indicates the start of re-epithelialization. Scale bar = 1mm. Tissues shown are representative of 2 independent experiments, each performed in triplicate.
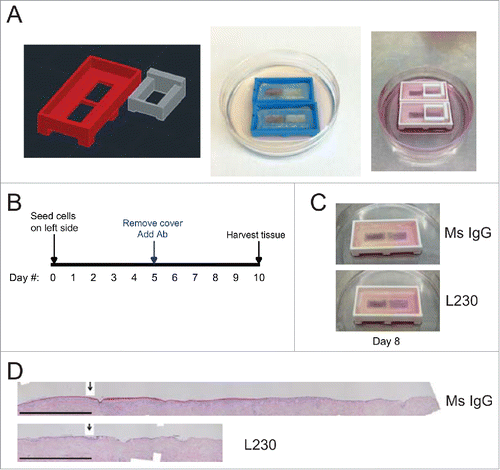
Figure 2. Integrin αv is necessary for proliferation during wound healing of human skin in vivo. (A) Representative Hematoxylin and Eosin (H&E) stains for wounds from mice treated with a control mouse IgG antibody or an L230 antibody. Wounds were harvested 2 days after wounding. B,C. Representative images of Ki67 and mouse secondary antibody labeling of wound adjacent tissue (B) or wound edge tissue (C). Dotted lines indicate the basement membrane zone. (D, E) Quantification of %Ki67+ epidermal cells (D) or epidermal thickness (E) at the wound edge and in wound adjacent tissue. Wound edge was considered within 0.25mm of wound. (F) Quantification of epidermal migration into the wound in mm. N = 4 mice per group for day 2 and 5 mice per group for day 4. NS = not statistically significant. P values were calculated using a student's t-test. Scale bar = 200 µm.
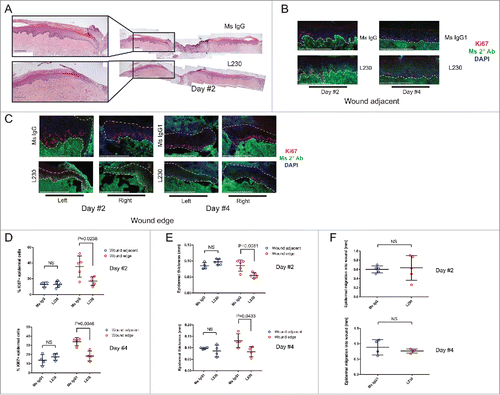
Figure 3. αv's role in re-epithelialization is partially dependent on TGFβ signaling. (A) Representative Hematoxylin & Eosin stain for runway re-epithelialization assay utilizing the indicated antibodies and TGFβ1 treatments. Arrows indicate the starting edge for the re-epithelialization assay. Scale bar= 1 mm. (B) Quantification of tissues from (A), in terms of epidermal thickness. Epidermal thickness was measured at various distances away from the starting edge of the re-epithelialization assay. N = 9 for Ms IgG, L230, and 10D5, N = 6 for L230 + 100 pM TGFβ and 10D5 + 100 pM TGFβ. p<0.005 (L230) and p<0.05 (10D5) by 2-way ANOVA. C. Quantification of cell layers from (A). Cell layers were counted at various distances away from the starting edge of the re-epithelialization assay. N = 9 for Ms IgG, L230, and 10D5, N = 6 for L230 + 100 pM TGFβ and 10D5 + 100 pM TGFβ. p < 0.001 (L230) and p < 0.001 (10D5) by 2-way ANOVA. D. Representative PCNA staining for tissues represented in panel (A). E. Quantification of the % of PCNA+ cells from panel (D) at the migratory edge. p < 0.05 using one-way ANOVA. *= p < 0.05, **= p < 0.005, and ***= p<0.0005 using Tukey's HSD post-hoc test. Scale bar = 100 µm.
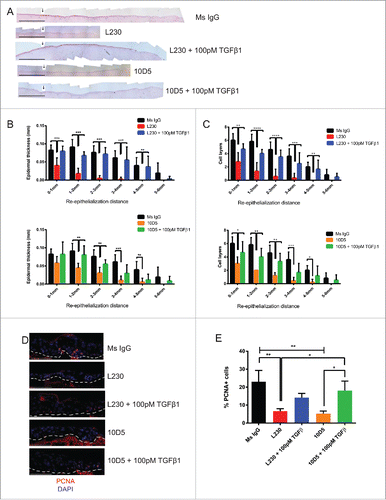
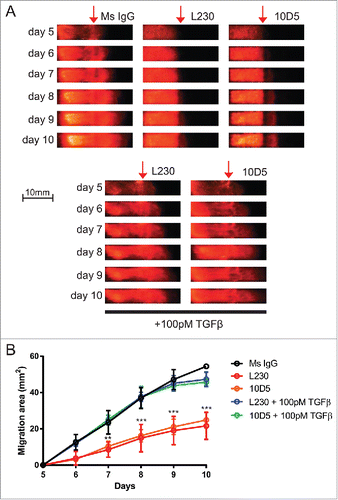
Figure 4. Epidermal migration is dependent on an αv-TGFβ signaling axis. (A) Representative fluorescent images of runway tissue seeded with keratinocytes labeled with a dsRed reporter. Tissues were treated with the indicated antibodies and recombinant proteins, which were added 5 days after seeding, and imaged every day. TGFβ1-containing media was replaced every day. (B) Quantification of the images in (A), normalized to the tissues at day 5, prior to treatments. N=3 tissues per group. p<0.0001 by 2-way ANOVA. **=p<0.005, ***=p<0.0005, calculated using Bonferroni post-hoc test. Comparisons are statistically significant between L230 vs. Ms IgG, L230 + 100 pM TGFβ and 10D5 + 100 pM TGFβ, and between 10D5 vs. Ms IgG, L230 + 100 pM TGFβ and 10D5 + 100 pM TGFβ.