Figures & data
Figure 1. Expression of BRCA1 wild type and missense variants in the strain RS112 of Saccharomyces cerevisiae. (A) Schematic representation of full length BRCA1 and the most relevant domains. Arrows indicate the missense variants we have studied. (B) Description of the BRCA1 missense variants; in the table, the localization domain, nucleotide and amino-acid change is reported. IARC classification is also listed; Class 5 highly pathogenic and Class 1 not pathogenic or neutral.Citation23 (C) BRCA1 was detected in the total protein extracts from yeast strains grown with galactose by Western blot analysis with anti BRCA1 antibody. Extracts were prepared from yeast expressing BRCA1wt, BRCA1-C61G, BRCA1-M1775R, BRCA1-mCherry, BRCA1- K45Q mCherry, BRCA1-C61G mCherry, BRCA1- R1751Q mCherry or BRCA1-M1775R mCherry, and loaded as indicated. Protein extract from yeast carrying empty pYES2 was loaded as control. In the lower part of the figure, the loading control was evaluated by detecting the 3PGK band is shown. The lower tables represent the densitometry calculated as ratio between intensity of BRCA1and PGK. Data are reported as mean of 3 independent experiments ± error bar. (D) RNA was exacted from yeast cells expressing BRCA1 wt, BRCA1-C61G, BRCA1-M1775R, BRCA1-mCherry, BRCA1- K45Q mCherry, BRCA1-C61G mCherry, BRCA1- R1751Q mCherry or BRCA1-M1775R mCherry, reverse transcribed and amplified for quantitative analysis as described in the Materials and Methods. Results are reported as mean of 4 experiments ± standard deviation.
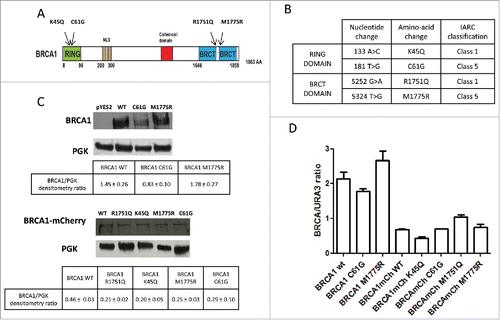
Table 1. Expression of BRCA1 wild type, BRCA1-C61G or BRCA1-M1775R decreases MMS resistance in the diploid strain RS112 of Saccharomyces cerevisiae. Yeast expressing BRCA1 were exposed to MMS in galactose medium for 17 hours; then, cells were washed counted and plated as described in Materials and Methods. Survival was determined as ratio between counted cells and number of colonies. Results are reported as mean of at least 4 experiments ± standard deviation. Statistics was carried out comparing results at the same MMS dose.a p ≤ 0.05; Statistical analysis was performed by comparing the survival of BRCA1 expressing cells to control (pYES2). * p ≤ 0.05, ** p ≤ 0.01; Statistical analysis was performed by comparing the survival of BRCA1-C61G and -M1775R expressing cells to that one of BRCA1wt expressing cells.
Table 2. Expression of BRCA1 wild type, BRCA1-C61G or BRCA1-M1775R affects MMS-induced intra-chromosomal recombination in the diploid strain RS112 of Saccharomyces cerevisiae. Yeast expressing BRCA1 were exposed to MMS in galactose medium for 17 hours; then, cells were washed counted and plated as described in Materials and Methods. Intra-chromosomal recombination was determined as total number of HIS3 colonies /104 survivors. Results are reported as mean of at least 4 experiments ± standard deviation. In the brackets, fold increase over the control (dose 0) is reported. Statistical analysis was carried out by comparing the frequency of MMS exposed cells to the control. * p ≤ 0.05, ** p ≤ 0.01, ***p ≤ 0.001.
Table 3. Expression of BRCA1 wild type, BRCA1-C61G or BRCA1-M1775R affects MMS-induced inter-chromosomal recombination in the diploid strain RS112 of Saccharomyces cerevisiae. Yeast expressing BRCA1 were exposed to MMS in galactose medium for 17 hours; then, cells were washed counted and plated as described in Materials and Methods. Intra-chromosomal recombination was determined as total number of ADE2 colonies /105 survivors. Results are reported as mean of at least 4 experiments ± standard deviation. In the brackets, fold increase over the control (dose 0) is reported. Statistical analysis was carried out by comparing the frequency of MMS exposed cells to the control. ** p ≤ 0.01, *** p ≤ 0.001.
Figure 2. Localization of BRCA1wt and missense variants in the strain RS112 of Saccharomyces cerevisiae after exposure with MMS. Yeast cells co-expressing BRCA1-mCherry fusion proteins and Nup133 were exposed to 100µg/ml MMS for 17 hours. Thereafter, cells were spotted onto glass slides and analyzed under the fluorescence microscope as described in the Materials and Methods. Merged images indicate that BRCA1 wt and all variants localized in the nucleus. In nontreated cells (control) BRCA1wt and all the missense variants localized mainly as “diffuse signal.” Some single nuclear focus was visible also in untreated cells expressing BRCA1 wt or missense variants; these single focused cells were not shown in the image because the number of cells with diffuse signal was more relevant; after MMS treatment, BRCA1wt and the neutral R1751Q localized as single nuclear focus (arrows).
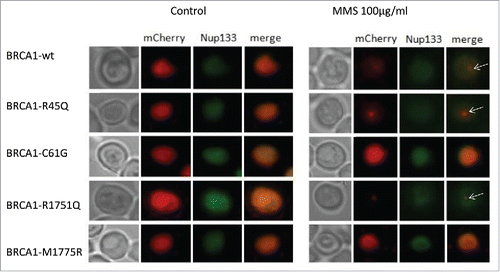
Table 4. Effect of MMS on yeast cells co-expressing BRCA1-mCherry fusion proteins. Cells were exposed to MMS as described in the Materials and Methods. Thereafter, cells were spotted onto glass slides and analyzed under the fluorescence microscope. Total red cells both with dispersed and focused localization were counted; in the bracket the cell number with a single focus was reported. Statistics was performed by “Chi-square” analysis on the real number of cells expressing each variant with or without MMS. Percentages, reported as mean ± standard deviation of 4 independent experiments, are shown for better comparison. * p ≤ 0.005, *** p ≤ 0.0001.
