Figures & data
Figure 1. The symmetrical distribution of PKH26-labeled subcellular compartments occurs in colorectal cancer stem cells. (A) The representative images for PKH26 dye labeling in HT29 cells at the indicated time points. PKH26 dye, red; DNA, blue. Scale bar = 20 μm. (B) The immunofluorescent pictures for showing the association of PKH26-labeled vesicles and endosome markers. PKH26 dye, red; endosome markers (EEA1, RAB11 and RAB7), green; DNA, blue. Scale bar = 5 μm. Arrows indicate the association or colocolization. Insert: magnified vesicle association indicated by yellow arrows. (C) The quantification results of association of PKH26-vesicles and indicated endosome markers in HT29 parental cells. Data represent mean ± SEM ***, p < 0.001 (Student's t test). (D) Representative images of symmetric or asymmetric segregation of PKH26-labeled vesicles in HT29 SDCSCs cultured under stem cell medium or in FBS-induced differentiation, respectively. PKH26 dye, red; DNA, blue. insert: phase pictures for showing paired-cells. (E) The percentage of the asymmetry/symmetry of PKH26-labeled vesicles in parental cells, SDCSCs and serum-differentiated SDCSCs (differentiation) in HT29 and HCT15 cells. n (total counted cells over 2 independent experiments) = 142, 223, 83, 144, 196, and 54 for HT29 parental cells, HT29 SDCSCs, Differentiation (HT29 SDCSCs), HCT15 parental cells, HCT15 SDCSCs, and Differentiation (HCT15 SDCSCs), respectively. PKH-Sym, symmetric segregation of PKH26-labeled vesicles; PKH-Asym, asymmetric segregation of PKH26-labeled vesicles. The p-value is estimated by χ2 test. *, p < 0.05; **, p < 0.01 ***, p < 0.001.
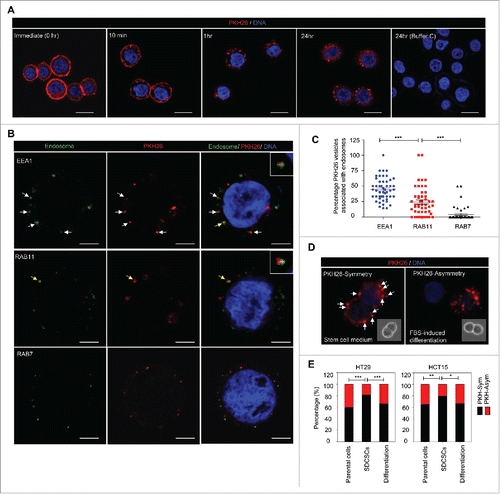
Figure 2. The PKH26 vesicles co-segregate into daughter stem cells divided from SDCSCs. (A) A schema for illustrating the paired-cell assay. IFA, immunofluorescence analysis. (B) Representative images of paired-cell assay of serum-differentiated HT29 SDCSCs. CD44, white; PKH26 dye, red. PKH-Sym, symmetric segregation of PKH26-labeled vesicles; PKH-Asym, asymmetric segregation of PKH26-labeled vesicles. (C) Left: The percentage of PKH26 vesicles symmetry/asymmetry in serum-differentiated HT29 SDCSCs. CD44 Sym, symmetric CD44 segregation; CD44 Asym, asymmetric CD44 segregation. n (total counted cells over 2 independent experiments) = 66 and 37 for CD44 Sym and CD44 Asym, respectively. ***, p < 0.001 (χ2 test). Right: The percentage of CD44 and PKH26 vesicles co-expression (CE) or inverse expression (IE) in PKHBright daughter cells undergoing asymmetric cell division. Data represent mean ± SD n = 3 independent experiments. *P < 0.05 (Student's t test). (D) Representative images of paired-cell assay of serum-differentiated HT29 SDCSCs. Snail, white; PKH26 dye, red. (E) Left: The percentage of PKH26 vesicles symmetry/asymmetry in serum-differentiated HT29 SDCSCs. Snail Sym, symmetric Snail segregation; Snail Asym, asymmetric Snail segregation. n (total counted cells over 2 independent experiments) = 87 and 38 for Snail Sym and Snail Asym, respectively. ***, p < 0.001 (χ2 test). Right: The percentage of Snail and PKH26 vesicles co-expression (CE) or inverse expression (IE) in PKHBright daughter cells undergoing asymmetric cell division. *, p < 0.05 (Student's t test). (F) Representative images of paired-cell assay of serum-differentiated HT29 SDCSCs. BMP4 and Numb, white; PKH26 dye, red. (G) The percentage of BMP4 and PKH26 vesicles co-expression (CE) or inverse expression (IE) in PKHDim daughter cells undergoing asymmetric cell division. (H) The percentage of Numb and PKH26 vesicles co-expression (CE) or inverse expression (IE) in PKHDim daughter cells undergoing asymmetric cell division.
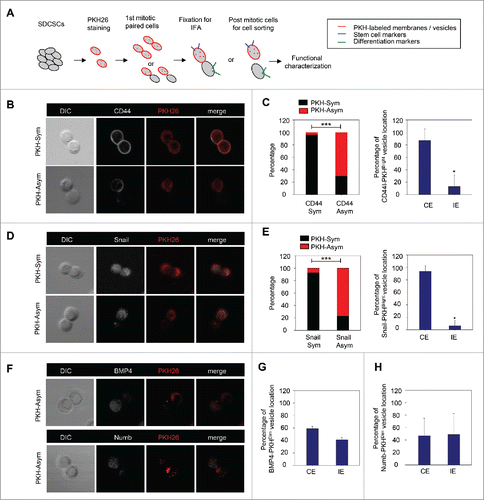
Figure 3. The PKHBright daughter stem cells exhibit a higher capability of forming tumorispheres and colonies. (A) Flow cytometry results for showing the gated regions of PKHBright (R1) and PKHDim (R2) of serum-differentiated HT29 SDCSCs before sorting and the purities after sorting. The percentage of cells in the gated region is illustrated in the corresponding panels. (B) Representative images of spheroids formed from HT29 SDCSCs- and HCT15 SDCSCs-derived PKHBright and PKHDim daughter cells. Scale bar = 100μm. (C) Histograms for showing the secondary sphere formation capacities of HT29 SDCSCs- and HCT15 SDCSCs-derived PKHBright and PKHDim daughter cells. Bright, PKHBright cells; Dim, PKHDim cells. Data represent mean ± SD n = 3 independent experiments. ***, p < 0.001 (Student's t test). (D) Histograms for showing the anchorage-independent colony forming capacities of HT29 SDCSCs- and HCT15 SDCSCs-derived PKHBright and PKHDim daughter cells. LPF, low power field. *, p < 0.05 (Student's t test).
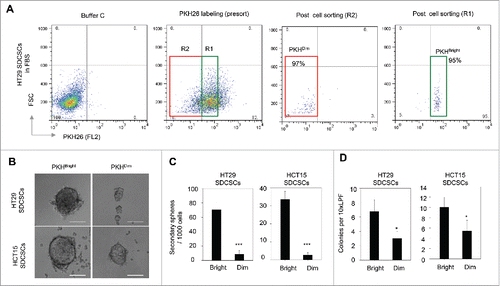
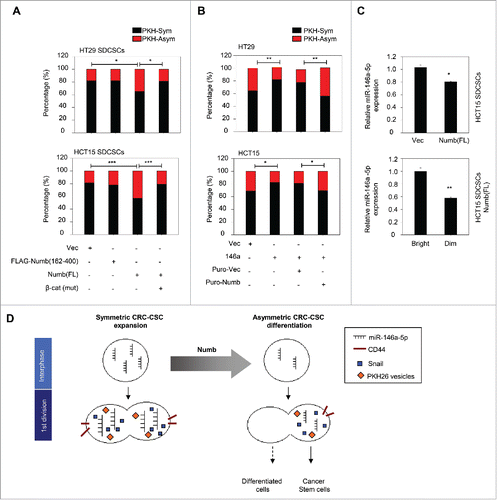
Figure 4. Restoration of Numb directs the asymmetry of PKH26 vesicles. (A) Histograms for showing percentages of PKH26 vesicle asymmetry/symmetry in the indicated stable cell lines. PKH-Asym, asymmetric segregation of PKH26 vesicles. n (total counted cells over 2 independent experiments) = 110, 34, 75, 123, 145, 137, 137, and 138 for HT29 SDCSC-Vec, HT29 SDCSCs-FLAG-Numb(162-400), HT29 SDCSCs-Numb(FL) and HT29 SDCSCs-Numb(FL)/β-cat(mut), HCT15 SDCSC-Vec, HCT15 SDCSCs-FLAG-Numb(162-400), HCT15 SDCSCs-Numb(FL) and HCT15 SDCSCs-Numb(FL)/β-cat(mut), respectively. The p-value is estimated by χ2 test. *, p < 0.05; ***, p < 0.001. (B) Histograms for showing percentages of PKH26 vesicle asymmetry/symmetry at indicated stable cell lines. n (total counted cells over 2 independent experiments) = 146, 148, 143, 147, 106, 75, 115 and 119 for HT29-Vec, HT29-146a, HT29-Puro-Vec, HT29-146a/Puro-Numb, HCT15-Vec, HCT15-146a, HCT15-Puro-Vec and HCT15-146a/Puro-Numb, respectively. The p-value is estimated by χ2 test. *, p < 0.05; **, p < 0.01. (C) Upper, RT-qPCR result for showing the expression of miR-146a-5p in HCT15 SDCSC-Vec and HCT15 SDCSC-Numb(FL) stable lines. Lower, RT-qPCR result for showing the expression of miR-146a-5p in PKHBright and PKHDim daughter cells of HCT15 SDCSCs-Numb(FL) cultured in stem-cell medium. PKHBright cells; Dim, PKHDim cells. *, p < 0.05; **, p < 0.01 (Student's t test). (D) A schema for summarizing the impact of Numb in switching division modes of colorectal cancer stem cells.