Figures & data
Figure 1. S100 (PDB ID: 1J55; rendered in dark green) shares a high degree of sequence and structural homology with Calmodulin (PDB ID: 3CLN; rendered in blue).
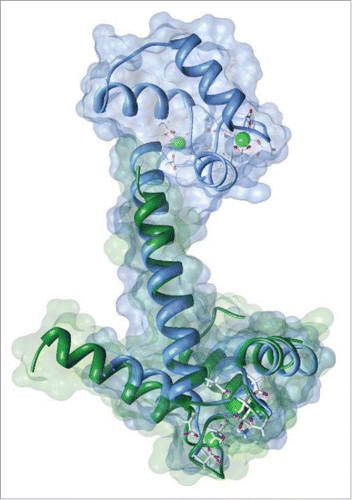
Figure 2. Correlation of HADDOCK scores with l-RMSD (ligand-root mean square-deviation) FCC(fraction of common contacts); i-l-RMSD (interface-ligand-RMSD for water refined structures) and i-RMSD (interface-RMSD). Top scoring cluster 2 (dark blue triangle) is circled in red.
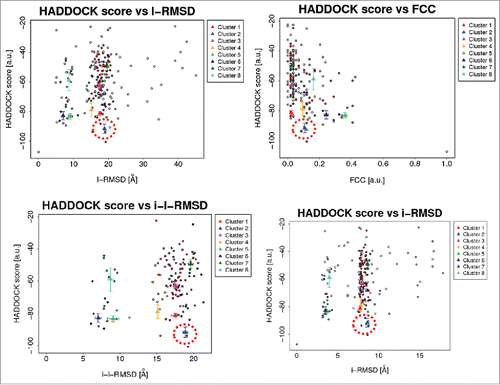
Figure 3. Docking pose of S100P –full length ERα complex. Atomic coordinate structure with surface representation of Cluster 2; S100P is rendered in green and ERα in pink.
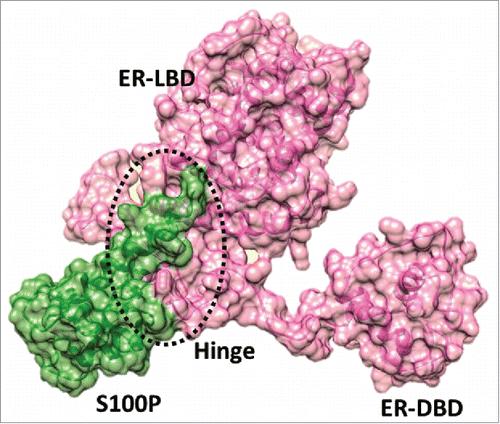
Table 1. Binding scores of the top 8 S100P-ERα complexes.
Figure 4. Binding scores of conformation-considered ER hinge peptide versus S100P. ER 298–317 helix/loop hinge peptide (gray star) scores were similar to that of ER full-length protein.
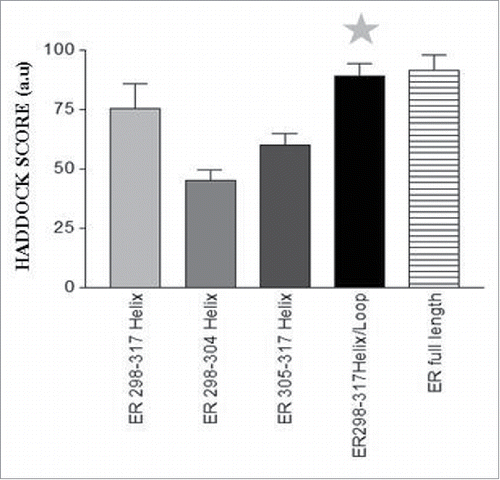
Table 2. Binding scores of ER hinge (conformation-considered peptides) -S100P complexes.
Figure 5. Active S100P residues that interact with estrogen receptor (pink) based on molecular modeling studies are highlighted in gray surfaces and green ribbons. S100P residues that reside within 5 A interaction zone is highlighted in yellow.
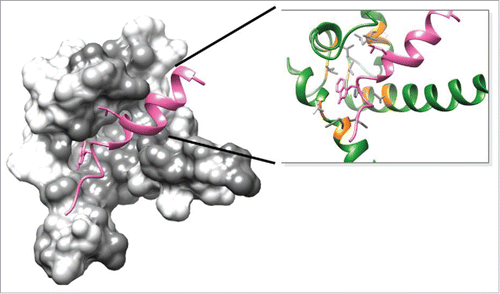
Figure 6. Docking pose of S100P (rendered in green) vs. ERα conformation considered hinge peptide (I298KRSKKNSLALSLTADQMV317S; rendered in pink). Atomic coordinate structure with surface representation of Cluster 2.
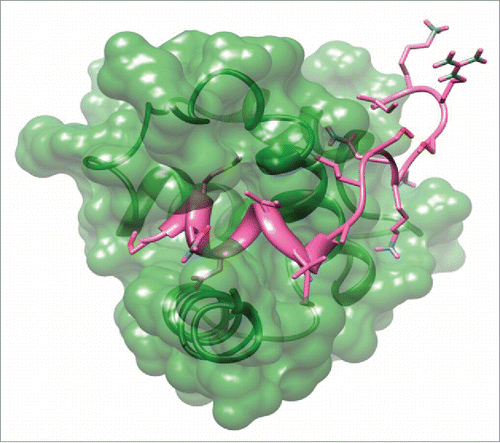
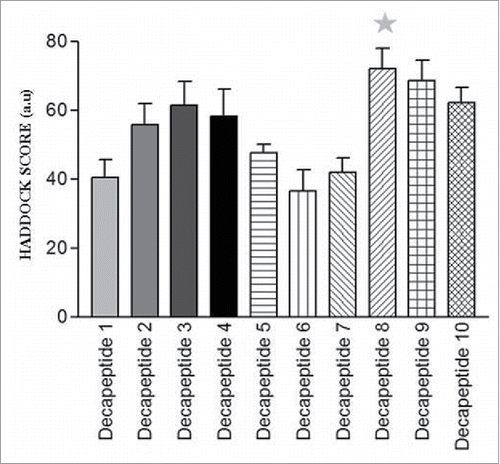
Table 3. Binding scores of top of S100P decapeptide-ERα complexes.
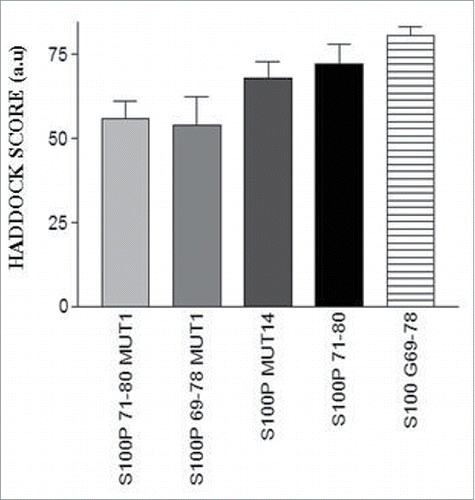
Table 4. Binding scores of short S100P mutant peptides-full length ERα complexes.
Figure 9. Representative images of the EdU cell proliferation assay done after MCF7 cells were subjected the following treatments (A) Vehicle (0.1% DMSO); (B) Estradiol 1nM; (C) S100 Peptide P1 5 uM and (D) S100 Peptide P1 5 uM + Estradiol 1 nM. All images were captured with a Zeiss Axio Observer inverted microscope at 10X magnification with filters appropriate for Hoechst and FITC fluorescence (Alexa Fluor 488- ex495nm, em519nm; NuclearMask Blue stain- ex350nm, em461nm). The total DNA content was stained with a NuclearMask Blue stain (blue), and EdU was visualized after conjugation with Alexa Fluor 488 azide (green).
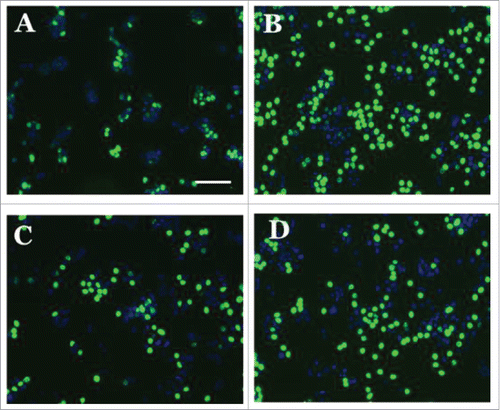
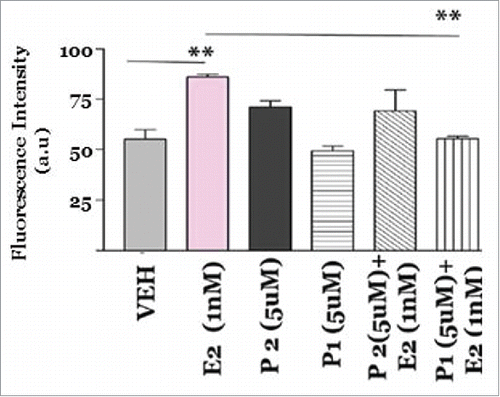
Figure 10. Effect of S100 peptides on MCF7 breast cancer cell proliferation. MCF7 cells were treated with A) Vehicle (0.1% DMSO); B) Estradiol 1nM; C) S100 Peptide P1 5 uM and D) S100 Peptide P1 5 uM + Estradiol 1 nM for 8 hours at 37oC with 5% CO2 and 10% humidity. A Click-iT 5-ethylnyl-2′-deoxyuridine (EDU) Alexa Fluor 488 based High Content Screening assay was performed in the 96 well format and fluorescence intensity was captured by employing 495nm excitation and 519nm emission filters with the Bio-Tek Synergy plate reader. All assays were run in triplicates. Error analysis and statistics was performed using GraphPad Prism. Results represent the mean ±standard deviation, where ** represents **p < 0.01.