Figures & data
Figure 1. PICH and UBF occupy different regions of the rDNA. PICH−/− cells ectopically expressing either hPICH (left) or hPICH-K128A (right) were arrested in prometaphase using nocodazole, released from this arrest, and then imaged at different stages of mitosis using structured illumination microscopy. Images were 3D rendered. PICH (red) and UBF (green) were detected using specific antibodies, and DNA (grayscale) was stained with DAPI. The small white boxes denote zoomed images of the minor NOR, and the large white boxes denote the major NORs. Scale bars, 5 μm.
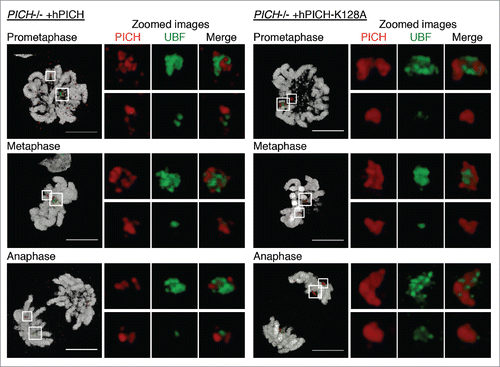
Figure 2. PICH does not appear to regulate rDNA transcription. (A) PICH−/− +hPICH cells were incubated with EU and imaged by immunofluorescence microscopy. EU (green) was detected using Alexa Fluor 488 azide, UBF (to denote the rDNA; red) was detected using a specific antibody, and the DNA (grayscale) was stained with DAPI. Scale bars, 5 μm. In the top panel, cells were arrested in G2 by RO3306 treatment for 6 hours before labeling with EU. In the middle panels, cells were released from the RO3306 arrest into medium containing nocodazole for 30 mins before labeling with EU. Cells in either prophase or prometaphase are shown. In the bottom panels, cells were arrested in prometaphase for 4.5 hours using nocodazole, and then released for 30 mins before labeling with EU. Cells in either metaphase or telophase are shown. (B/C) Asynchronously growing wild-type and PICH−/− cells were incubated with EU for 15 mins, and then imaged by immunofluorescence microscopy. rDNA transcription around the major DT40 rDNA locus was quantified in prometaphase and in late telophase cells (panel B) as described in the Materials and Methods. More than 100 prometaphase or late telophase cells were quantified in 3 independent experiments. Data points are averages ± s.d. (C) Representative examples of wild-type cells in prometaphase and late telophase stained for EU (red) and UBF (green). DNA (grayscale) was stained with DAPI. Scale bars, 5 μm.
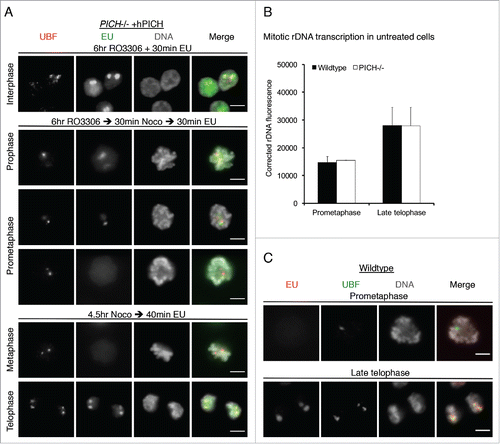
Figure 3. The rDNA marks a novel class of UFBs. (A,C-E) PICH−/− cells ectopically expressing either hPICH or hPICH-K128A were arrested in prometaphase using nocodazole then released into drug-free medium or medium containing 0.25 μM ICRF-193 for 40 min, as indicated. (A-C) PICH (red) and UBF (green) were detected using specific antibodies and the DNA (grayscale) was stained with DAPI. Scale bars, 5 μm. In panel A, cells were imaged using structured illumination microscopy and the images were 3D rendered. White boxes denote zoomed images. (B) An example of a DAPI-negative rDNA-UFB in an untreated PICH−/− cell. (C) Representative examples of rDNA-UFBs arising from either the major or the minor rDNA locus in PICH deficient cells expressing either wild-type hPICH or hPICH-K128A (as indicated). (D/E) Quantification of total UFBs per anaphase (left), of rDNA-UFBs/anaphase (middle), and of rDNA-UFBs/UFB (right) in PICH deficient cells expressing either wild-type hPICH or hPICH-K128A (as indicated). Each data point is an average of at least 3 independent experiments. Significance levels were calculated using a Students t-test for parametric observations and are indicated as *P < 0.05, **P < 0.01 and ***P < 0.001.
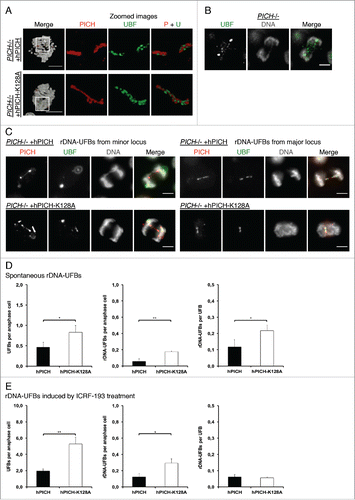
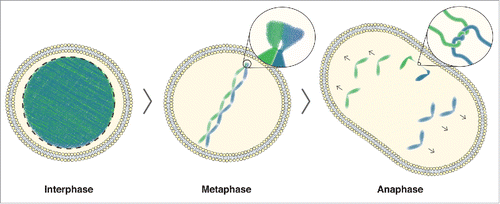
Figure 4. rDNA-UFB model. In interphase cells, DNA is uncondensed. As cells enter mitosis, the DNA begins to condense, with the exception of the rDNA. In metaphase cells, the condensed chromosomes are aligned at the equator of the cell. The zoomed image shows the rDNA where delayed condensation causes a delay in decatenation of any DNA entanglement that arose due to DNA replication or homologous recombination. In anaphase, this can lead to DNA bridging, as shown in the zoomed image of a catenated DNA bridge. Ongoing condensation, in combination with the tension building up on unresolved catenanes by the spindle pulling forces, allows efficient decatenation of any rDNA-UFBs during anaphase.