Figures & data
Figure 1. Cbf11 regulates gene expression in an apparently cell-cycle phase-specific manner after cdc25–22 block-release. (A) Timecourse ChIP analysis of Cbf11 binding to cut6, vht1 and bio2 promoters and control unbound loci in cells synchronized in late G2 by cdc25–22 block-release. S phase coincides with division septum formation (see ‘septation index’). Results representative of 2 independent experiments are shown. (B) Comparison of normalized Cbf11 ChIP-seq signal (area under ChIP-seq peak) from unsynchronizedCitation26 and S-phase/cytokinesis cells synchronized by cdc25–22 block-release. Pooled data from 2 independent experiments are shown. (C) Timecourse RT-qPCR analysis of cut6, vht1 and bio2 mRNA levels in wild-type and Δcbf11 cells synchronized by cdc25–22 block-release. The cdc22 gene is a positive control, peaking at the G1/S boundary.Citation32 Results representative of 2 independent experiments are shown.
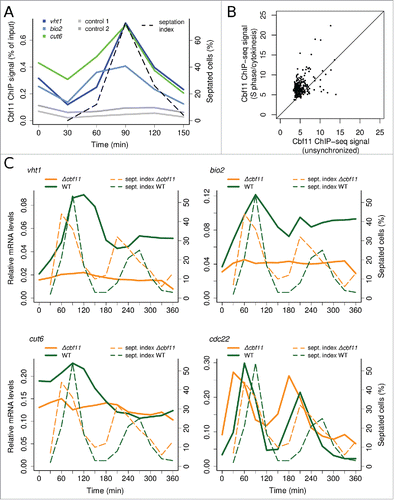
Figure 2. Genes with markedly increased Cbf11 binding to their promoters during S phase/cytokinesis following cdc25–22 block-release. (A) Pooled data from 2 independent ChIP-seq experiments are shown. ‘n/s’ - not synchronized, ‘S’ - S phase/cytokinesis. (B) Average expression levels of genes from (A) during 2 cell cycles, post cdc25–22 block-release. Data were taken from ref. 7. Expression peaks around S/G2.
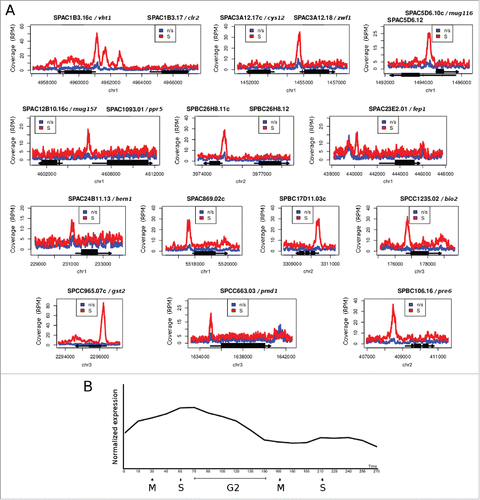
Figure 3. Expression patterns of Cbf11 target genes depend on synchronization method. Timecourse RT-qPCR analysis of cut6, vht1 and bio2 mRNA levels in wild-type cells synchronized by cdc10-M17 block-release (A), hydroxyurea block-release (B), and lactose gradient centrifugation (C). S phase coincides with division septum formation (see ‘septation index’); note that for the synchronization method used in panel (A) the very first S phase is never accompanied by septation. The cdc22 gene is a positive control, peaking at the G1/S boundary.Citation32 Note that cdc22 is induced by hydroxyurea treatment even at time 0 in (B).Citation7 Results representative of 3 independent experiments are shown.
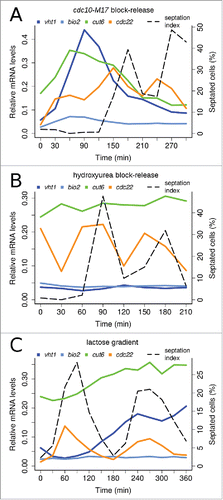
Figure 4. Cbf11 is a direct activator of cut6 expression. (A) RT-qPCR analysis of cut6 expression. Mean values ± SD from 3 independent experiments are shown. (B) Differential occurrence of catastrophic mitosis in cells grown in YES and EMM. Mean values ± SD from 6 independent experiments are shown. Significance was determined by one-sided paired t-test (**p ≤ 0.01; ‘n.s.’ p > 0.05). (C) Deletion of cbf11 shows a severe synthetic sick genetic interaction with cut6–621 at permissive temperature. Segregants from 3 representative meioses are shown. (D) The Δcbf11 strain, but not the Pcut6MUT promoter mutant strain, showed reduced growth compared to wild type. Results representative of ≥3 independent experiments are shown. (D) Effect of cut6 overexpression on wild-type and Δcbf11 cell proliferation. Results representative of 3 independent experiments are shown.
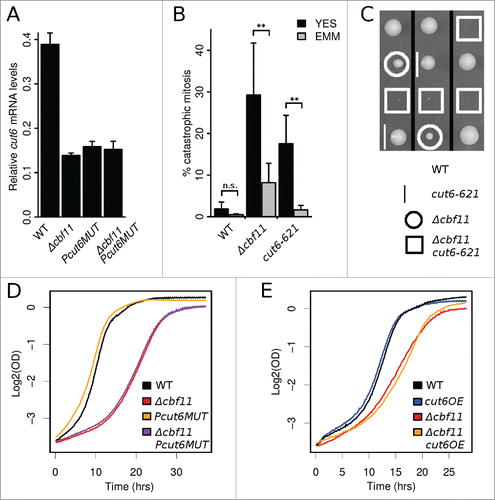
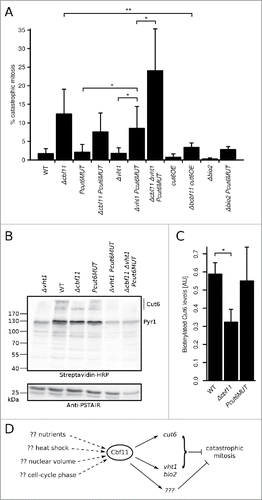
Figure 5. Cbf11-regulated genes are needed to prevent catastrophic mitosis. (A) Cells were fixed, stained with DAPI and the occurrence of catastrophic mitosis was determined. Mean values ± SD from ≥3 independent experiments are shown. Significance was determined by one-sided unpaired t-test (*p ≤ 0.05; **p ≤ 0.01). (B) Western blots of whole cell extracts were probed with streptavidin-HRP to visualize biotinylated proteins, and with anti-PSTAIR antibody as a loading control. The position of Cut6 (2 bands) and the expected position of pyruvate carboxylase Pyr1 are indicated. Results representative of 3 independent experiments are shown. (C) Densitometric quantification of Cut6 ECL signals from (B). Mean values ± SD of 3 independent repeats are shown. Significance was determined by one-sided paired t-test (*p ≤ 0.05). (D) Model of gene expression regulation by the Cbf11 transcription factor. Cbf11 binds to target promoters and activates transcription of genes required to prevent catastrophic mitosis. Speculative inputs influencing Cbf11 activity are indicated on the left and discussed in the main text.