Figures & data
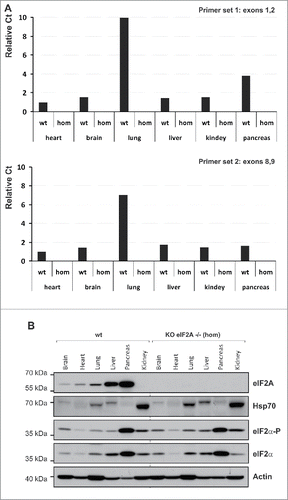
Figure 1. The eIF2A gene. (A) Top: Mus musculus eukaryotic translation initiation factor 2A (eIF2A) (mouse Accession: NM_001005509) gene organization and the site of insertion of Omnibank Vector 74. Bottom: Mouse genomic sequence surrounding the gene trap insertion site identified in the C57BL/6 gene trap ES cell clone IST13504C3sA4 for eIF2A. The initiating ATG codon is in red and the coding part of exon 1 is in cyan. The insertion site is denoted with an asterisk *. (B) Top: Schematic of the insertion site and the genotyping strategy. Relative positions of the primers and the expected sizes of the PCR fragments are indicated. Bottom: Genotyping results (1.5% agarose gel).
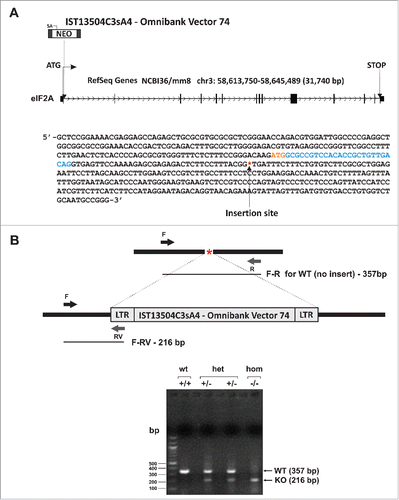
Figure 2. The eIF2A gene expression. (A) Real-time PCR. Relative Ct values are shown; Ct values were normalized to gapdh signal and are shown relative to wild-type brain sample. Two sets of primers were used: Primer set 1, recognizing sequences within exons 1 and 2 (top) and primer set 2 recognizing sequences within exons 8 and 9 (bottom). (B) Western blotting. mRNA and protein levels were analyzed in 6 different mouse tissues heart, brain, lung, liver, kidney and pancreas, respectively.