Figures & data
Figure 1. Atd1, Atd2 and Atd2 are involved in acetaldehyde detoxification. (A, B) Five-fold serial dilutions of cells of the indicated genotypes were grown on YES agar medium containing 0, 15, and 25 mM acetaldehyde. Cells were incubated for 3 to 5 d at 30°C. As described in the methods section, due to the difficulty in preventing acetaldehyde evaporation, the initial concentrations of acetaldehyde in the medium are shown. Representative images of repeat experiments are shown.
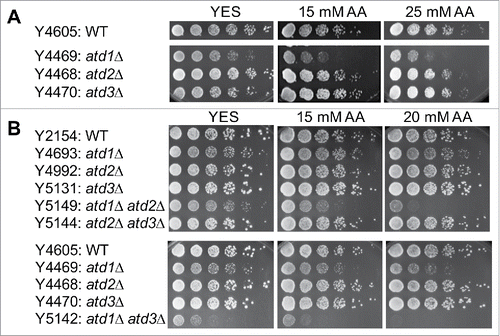
Figure 2. Acetaldehyde induces DNA damage in the absence of acetaldehyde dehydrogenase. (A) Cells were incubated for 3 h in YES liquid medium containing the indicated concentrations of acetaldehyde. Then, 5-fold serial dilutions of cells were prepared, plated on YES agar medium without acetaldehyde, and incubated for 3 to 5 d at 30°C. (B) Cells of the indicated genotypes were engineered to express Rad52-YFP, grown in YES medium at 25°C until mid-log phase, and treated with 10 mM acetaldehyde (AA) for 2 h. The percentages of nuclei with at least one Rad52-YFP focus are shown. At least 200 cells were counted for each strain. Error bars correspond to standard errors of the mean (SEM) obtained from 3 independent experiments. (C) Representative images of the acetaldehyde-treated cells used in B are shown. (D) Five-fold serial dilutions of cells of the indicated genotypes were grown on YES agar medium containing 0 and 20 mM acetaldehyde. Cells were incubated for 3 to 5 d at 30°C.
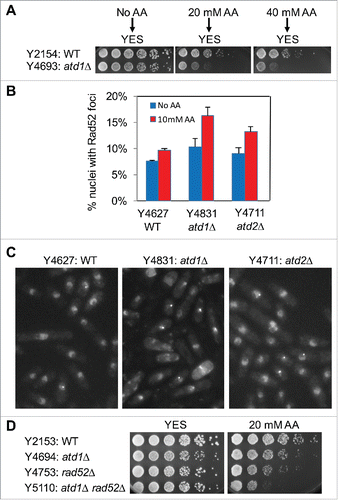
Figure 3. Acetaldehyde causes replication stress that activates the ATR-dependent checkpoint. (A, B) Five-fold serial dilutions of the indicated mutants were incubated on YES agar medium supplemented with the indicated drugs for 3 to 5 d at 30°C. Representative images of repeat experiments are shown. (C) PFGE analysis of chromosome samples prepared from wild-type and swi1Δ cells. Cells were grown in the presence of 20 mM acetaldehyde for 3 h and returned to fresh medium. Cells were collected at 0 (AA), 2, and 4 h after acetaldehyde treatment and processed for PFGE. Chromosomes prepared from exponentially growing cells are also analyzed (Log). Representative results of repeat experiments are shown. swi1Δ cells are known to display shorter chromosome III due to the elevated recombination rate at rDNA repeatsCitation50,60.
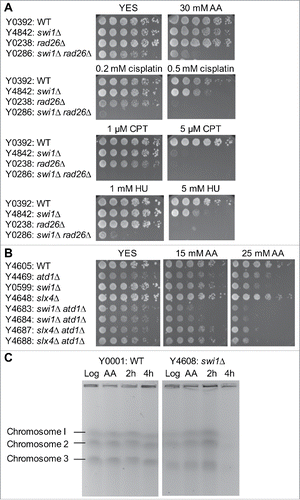
Figure 4. NER and HR play critical roles in repair of acetaldehyde-induced DNA damage. (A, B) Five-fold serial dilutions of the indicated mutants were incubated on YES agar medium supplemented with the indicated drugs for 3 to 5 d at 30°C. NER and HR mutants show strong sensitivity to acetaldehyde. Representative images of repeat experiments are shown. (C, D) Rad52-YFP foci analysis of the indicated cells was performed as described in . The percentages of nuclei with at least one Rad52-YFP focus and 2 or more foci are shown. Representative images of repeat experiments are shown.
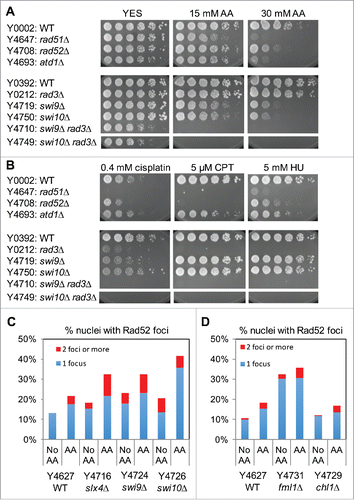
Figure 5. Wss1-like proteases are involved in cellular tolerance to acetaldehyde. Five-fold serial dilutions of the indicated mutants were incubated on YES agar medium supplemented with the indicated amounts of acetaldehyde (AA) for 3 to 5 d at 30°C. wss1Δ and swi1Δ have additive genetic interaction in acetaldehyde sensitivity. Representative images of repeat experiments are shown.
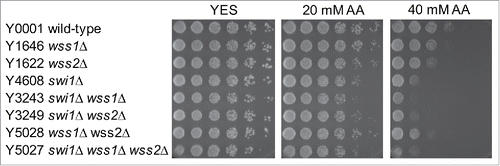
Figure 6. Involvement of the FA pathway in acetaldehyde-mediated DNA damage response. Five-fold serial dilutions of the indicated mutants were incubated on YES agar medium supplemented with the indicated drugs for 3 to 5 d at 30°C. (A, B, C) FA mutants show significant acetaldehyde sensitivity when combined with rad3Δ. (D) pso2Δ and fan1Δ nuclease mutants failed to show significant acetaldehyde sensitivity. Representative images of repeat experiments are shown.
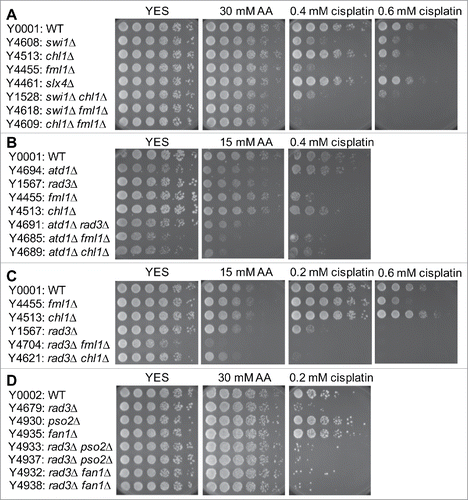
Figure 7. Epistasis analysis of FA, NER, HR and swi1Δ mutants. Five-fold serial dilutions of the indicated mutants were incubated on YES agar medium supplemented with the indicated drugs for 3 to 5 d at 30°C. (A) fml1Δ shows epistatic interaction with swi9Δ but not with swi1Δ. (B) fml1Δ has additive or synergistic genetic interaction with rad52Δ. Representative images of repeat experiments are shown.
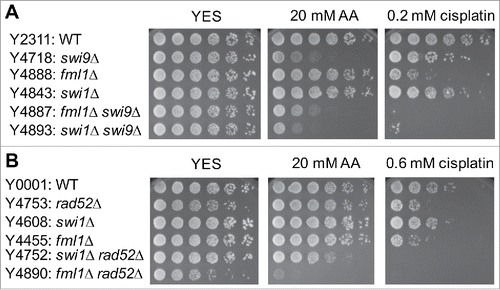
Figure 8. Involvement of TLS and BER in acetaldehyde-mediated DNA damage response. Five-fold serial dilutions of the indicated mutants were incubated on YES agar medium supplemented with the indicated drugs for 3 to 5 d at 30°C. (A) Single TLS mutants failed to show acetaldehyde and cisplatin sensitivity. (B) TLS polymerases become more important for the cellular tolerance to acetaldehyde when Rad3 is absent. (C) BER is involved in acetaldehyde-mediated DNA damage response. Representative images of repeat experiments are shown.