Figures & data
Figure 1. Structures of Brr2-Jab1 complexes. (A) Schematics of the Brr2 constructs used in the present and former studies. N-terminal truncations (T1, T2, T3 and T4; start sites indicated by angled arrows) were combined with deletion of the CC (in parentheses) in some cBrr2 variants investigated. (B) Diametric views of the yBrr2FL-Jab1 complex. (C) Diametric views of the yBrr2T2-Jab1 complex. (D) Diametric views of the cBrr2T3-Jab1 complex. (E) Diametric views of the hBrr2T3-Jab1 complex (PDB ID 4KIT).Citation23 Domains and regions are colored identically in this and all following figures. NTR – magenta; RecA1 – light gray; RecA2 – dark gray; WH – black; HB – blue; HLH – red; IG – green; CC – beige; Jab1 – gold. Regions of the NTRs and Jab1 domains present in the various structures are highlighted by semi-transparent surfaces. Circles in (B-E) – residues of the NC-clamp (436–443 in yBrr2, 418–425 in hBrr2, 467–474 in cBrr2) not defined in the electron densities of the yBrr2FL-Jab1 (B) and yBrr2FL-Jab1ΔC Citation20 structures. Structures were aligned according to their Jab1 domains.
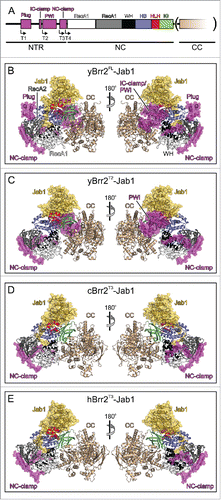
Table 1. Crystallographic data.
Figure 2. Interactions of the Jab1 C-terminal tail with the RNA-binding tunnel of Brr2. (A) Sequence alignment of the Jab1 tails of S. cerevisiae, C. thermophilum and H. sapiens. Residues interacting at the RNA-binding tunnel of the respective Brr2 protein are indicated by asterisks, colored according to the respective interacting Brr2 domain (RecA1 – light gray; RecA2 – dark gray; HB – blue; HLH – red). Residues affected by prp8 mutations that cause retinitis pigmentosa in humans are indicated by cyan dots above the alignment. Regions corresponding to the proximal, central and distal portions of the tails are indicated below the alignment. (B-D) Details of the interactions of the Jab1 C-terminal tails at the Brr2 RNA-binding tunnels in the yBrr2FL-Jab1 complex (B), cBrr2T3-Jab1 complex (C) and hBrr2T3-Jab1 complex (PDB ID 4KIT)Citation23 (D). Top panels – interactions of the central regions of the Jab1 tails. Bottom panels – interactions of the distal regions of the Jab1 tails. Interacting residues are shown as sticks and colored by atom type (carbon – as the respective domain/region; nitrogen – blue; oxygen – red). For interactions involving only protein backbone atoms, side chains are not shown for clarity. Dashed lines indicate hydrogen bonds or salt bridges. Rotation symbols indicate orientations relative to respectively.
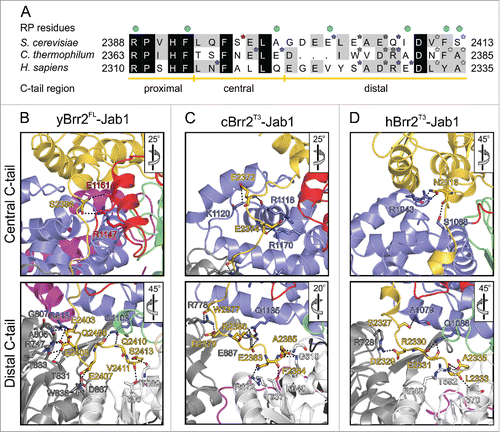
Table 2. Contacts between the Jab1 C-terminal tail and the Brr2 N-terminal cassette.
Figure 3. U4/U6 binding by Brr2 and possible inhibitory mechanisms. (A) yBrr2-Jab1 bound to U4/U6 as seen in the cryo-EM structure of a yeast U4/U6•U5 tri-snRNP (PDB ID 5GAO).Citation27 (B) Structure of the isolated yBrr2FL-Jab1 complex with portions of U4/U6, obtained by superimposition with the structure in (A) according to the NCs of the Brr2 subunits. (C) The NC-clamp and part of the PWI domain of the NTR bind U4/U6 stem II in the yeast U4/U6•U5 tri-snRNP. (D) The HLH domain of the NC contacts the U4 3′-SL in the yeast U4/U6•U5 tri-snRNP. (E) In isolated yBrr2FL-Jab1, the plug domain of the NTR sterically hinders accommodation of the stem I portion of U4/U6 between the RecA2 and HB domains of the NC. (F) In isolated yBrr2FL-Jab1, the Jab1 C-terminal tail binds along and hinders opening of the HB and RecA2 domains to accommodate RNA and occupies part of the tunnel that accommodates the ss region of U4 snRNA neighboring U4/U6 stem I during U4/U6 unwinding. U4 snRNA – brown; U6 snRNA – orange.
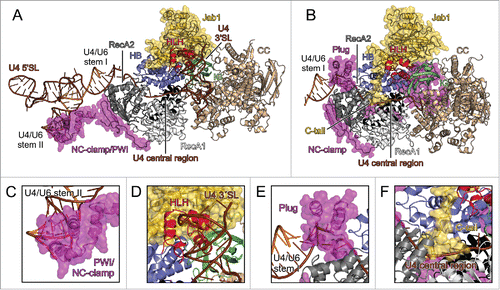
Figure 4. Effects of the NTR and the Prp8 Jab1 domain on RNA binding and unwinding by Brr2. (A,B) Assays monitoring RNA binding by hBrr2FL (A) or hBrr2T3 (B). Black curves – hBrr2FL or hBrr2T3 alone; gray curves – hBrr2FL or hBrr2T3 in complex with Jab1; red curves – hBrr2FL or hBrr2T3 in complex with JabΔC. Left panels – binding to ssRNA; central panels – binding to dsRNA; right panels – binding to yU4/U6 snRNA. Binding of ssRNA and dsRNA was monitored by FP using 5-FAM-labeled RNAs. Binding of yU4/U6 was monitored via EMSA. Data points and error bars represent means +/− SEMs of at least 2 independent experiments. (C) RNA affinities. Values represent means +/− SEMs of at least 2 independent experiments. Kd values were obtained by fitting the data from (A) and (B) to a single exponential Hill function (fraction bound = A[protein]n/([protein]n+Kdn); A – fitted maximum of RNA bound; n – Hill coefficient).Citation50 (D) Scheme summarizing the effects of the NTR and the Jab1 domain on different portions of a substrate. Red symbols – inhibition; green double-arrow – reinforcement. (E) U4/U6 unwinding by Brr2 variants in the absence or presence of Jab1 variants. Black curves – Brr2FL or Brr2T3 alone; gray curves – Brr2FL or Brr2T3 in complex with Jab1; red curves – Brr2FL or Brr2T3 in complex with JabΔC. (F) Unwinding parameters. Amplitudes and unwinding rates were obtained by fitting the data to a first-order reaction (fraction unwound = A[1-exp{-ku t}]; A – amplitude of the reaction; ku – apparent first-order rate constant of unwinding; t – time). Values represent means +/− SEMs of at least 4 independent experiments.
![Figure 4. Effects of the NTR and the Prp8 Jab1 domain on RNA binding and unwinding by Brr2. (A,B) Assays monitoring RNA binding by hBrr2FL (A) or hBrr2T3 (B). Black curves – hBrr2FL or hBrr2T3 alone; gray curves – hBrr2FL or hBrr2T3 in complex with Jab1; red curves – hBrr2FL or hBrr2T3 in complex with JabΔC. Left panels – binding to ssRNA; central panels – binding to dsRNA; right panels – binding to yU4/U6 snRNA. Binding of ssRNA and dsRNA was monitored by FP using 5-FAM-labeled RNAs. Binding of yU4/U6 was monitored via EMSA. Data points and error bars represent means +/− SEMs of at least 2 independent experiments. (C) RNA affinities. Values represent means +/− SEMs of at least 2 independent experiments. Kd values were obtained by fitting the data from (A) and (B) to a single exponential Hill function (fraction bound = A[protein]n/([protein]n+Kdn); A – fitted maximum of RNA bound; n – Hill coefficient).Citation50 (D) Scheme summarizing the effects of the NTR and the Jab1 domain on different portions of a substrate. Red symbols – inhibition; green double-arrow – reinforcement. (E) U4/U6 unwinding by Brr2 variants in the absence or presence of Jab1 variants. Black curves – Brr2FL or Brr2T3 alone; gray curves – Brr2FL or Brr2T3 in complex with Jab1; red curves – Brr2FL or Brr2T3 in complex with JabΔC. (F) Unwinding parameters. Amplitudes and unwinding rates were obtained by fitting the data to a first-order reaction (fraction unwound = A[1-exp{-ku t}]; A – amplitude of the reaction; ku – apparent first-order rate constant of unwinding; t – time). Values represent means +/− SEMs of at least 4 independent experiments.](/cms/asset/c2d6f72c-d3d1-47e4-861f-a5385d6074c2/kccy_a_1255384_f0004_c.gif)
Figure 5. Effects of NTR truncations on C. thermophilum Brr2NC variants. (A) Binding of N-terminal Brr2NC truncations to U4/U6 di-snRNA monitored by electrophoretic mobility shift assays. Values represent means ± SEMs of at least 2 independent experiments. Kd values were obtained by fitting quantified binding data to a single exponential Hill function (fraction bound = A[protein]n/([protein]n+Kdn); A – fitted maximum of RNA bound; n – Hill coefficient). (B) Time courses of U4/U6 unwinding by Brr2NC variants. Data represent means ± SEM of at least 3 independent experiments. Amplitudes and unwinding rates were obtained by fitting the quantified data to a first-order reaction (fraction unwound = A[1-exp{-ku t}]; A – amplitude of the reaction; ku – apparent first-order rate constant of unwinding; t – time). (C) U4/U6 di-snRNA affinities and unwinding parameters of Brr2NC variants. Values represent means ± SEM of at least 2 independent experiments.
![Figure 5. Effects of NTR truncations on C. thermophilum Brr2NC variants. (A) Binding of N-terminal Brr2NC truncations to U4/U6 di-snRNA monitored by electrophoretic mobility shift assays. Values represent means ± SEMs of at least 2 independent experiments. Kd values were obtained by fitting quantified binding data to a single exponential Hill function (fraction bound = A[protein]n/([protein]n+Kdn); A – fitted maximum of RNA bound; n – Hill coefficient). (B) Time courses of U4/U6 unwinding by Brr2NC variants. Data represent means ± SEM of at least 3 independent experiments. Amplitudes and unwinding rates were obtained by fitting the quantified data to a first-order reaction (fraction unwound = A[1-exp{-ku t}]; A – amplitude of the reaction; ku – apparent first-order rate constant of unwinding; t – time). (C) U4/U6 di-snRNA affinities and unwinding parameters of Brr2NC variants. Values represent means ± SEM of at least 2 independent experiments.](/cms/asset/3dfc1863-a0f4-4d32-824b-6a46091d2cca/kccy_a_1255384_f0005_b.gif)
