Figures & data
Figure 1. OSM/STAT3-induced senescence requires TGF-β signaling. (A) Relative growth assays (top) and western analysis of whole cell lysates (bottom) of shp53/vector-, DN-STAT3-, or DN-TGFβR2-HMEC plated in the presence (+) and absence (-) of recombinant OSM [10 ng/mL] or TGF-β1 [5 ng/mL] for 7 d. (B) Relative growth assays (top) and western analysis (bottom) of shp53-HMEC treated with recombinant OSM alone or in combination with one of 2 different small molecule TGFβR1 inhibitors, SB525334 [1 μM] (left) and SB431542 [5 μM] (right) for 7 d.
![Figure 1. OSM/STAT3-induced senescence requires TGF-β signaling. (A) Relative growth assays (top) and western analysis of whole cell lysates (bottom) of shp53/vector-, DN-STAT3-, or DN-TGFβR2-HMEC plated in the presence (+) and absence (-) of recombinant OSM [10 ng/mL] or TGF-β1 [5 ng/mL] for 7 d. (B) Relative growth assays (top) and western analysis (bottom) of shp53-HMEC treated with recombinant OSM alone or in combination with one of 2 different small molecule TGFβR1 inhibitors, SB525334 [1 μM] (left) and SB431542 [5 μM] (right) for 7 d.](/cms/asset/68af2cf1-2706-49d0-93e7-c3fc489a845c/kccy_a_1259037_f0001_b.gif)
Figure 2. OSM/STAT3-induced senescence requires SMAD3/SMAD4. (A) Schematic of canonical TGF-β/SMAD signaling-induced senescence in normal HMEC. (B) Relative growth assays (top) and Western analysis (bottom) of shp53-HMEC expressing either empty vector, DN-STAT3, DN-TGFβR2, SMAD7, or a constitutively-active Cyclin D1/CDK2 fusion protein (CA-D1/CDK2) plated in the presence (+) and absence (-) of recombinant OSM for 7 d. (C) Relative growth assays (left) and western analysis (right) of shp53-HMEC stably expressing shRNAs targeting either green fluorescent protein (shGFP), SMAD2 (shSMAD2), SMAD3 (shSMAD3), or SMAD4 (shSMAD4) plated in the presence (+) and absence (-) of recombinant OSM for 7 d.
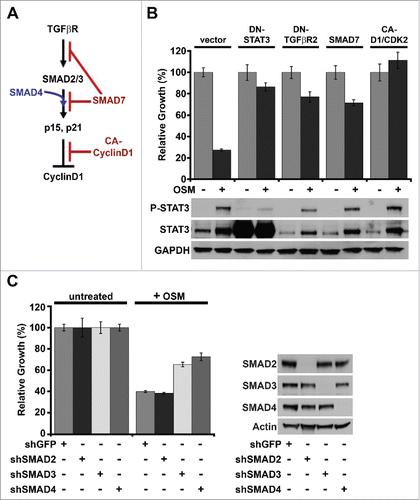
Figure 3. OSM/STAT3 signaling promotes SMAD3 nuclear localization without increased phosphorylation. (A) Western analysis of whole cell lysates harvested from shp53-HMEC plated in the presence (+) and absence (-) of either OSM or TGF-β1 for 4 and 8 d. (B) Western analysis of the nuclear-soluble protein fractions from subcellular fractionations of untreated shp53-HMEC and shp53-HMEC treated with either OSM or TGF-β1 for 4 d. (C) Western analysis of SMAD3-specific immunoprecipitations using whole cell lysates from both untreated and OSM-treated HEK 293T cells transfected with an expression vector encoding SMAD3 in combination with either an expression vector encoding wild-type STAT3 (WT-STAT3) or encoding dominant-negative STAT3 (DN-STAT3), and then plated in the presence (+) and absence (-) of recombinant OSM [10 ng/mL] for 2 d.
![Figure 3. OSM/STAT3 signaling promotes SMAD3 nuclear localization without increased phosphorylation. (A) Western analysis of whole cell lysates harvested from shp53-HMEC plated in the presence (+) and absence (-) of either OSM or TGF-β1 for 4 and 8 d. (B) Western analysis of the nuclear-soluble protein fractions from subcellular fractionations of untreated shp53-HMEC and shp53-HMEC treated with either OSM or TGF-β1 for 4 d. (C) Western analysis of SMAD3-specific immunoprecipitations using whole cell lysates from both untreated and OSM-treated HEK 293T cells transfected with an expression vector encoding SMAD3 in combination with either an expression vector encoding wild-type STAT3 (WT-STAT3) or encoding dominant-negative STAT3 (DN-STAT3), and then plated in the presence (+) and absence (-) of recombinant OSM [10 ng/mL] for 2 d.](/cms/asset/3e9026fb-8960-45e1-9078-12236e717003/kccy_a_1259037_f0003_b.gif)
Figure 4. Persistent OSM/STAT3 signaling promotes SMAD target-gene transcription. (A) mRNA harvested from shp53-HMEC plated in the presence (+) and absence (-) of either recombinant OSM [10 ng/mL] or recombinant TGF-β1 [5 ng/mL] for 7 d was subjected to a targeted TGF-β Signaling-Targets qRT-PCR profiler array. Genes exhibiting a similar change in expression at least 2-fold following treatment with both OSM and TGF-β1 were selected and displayed to compare gene expression between untreated shp53-HMEC and shp53-HMEC treated with either OSM or TGF-β1. (B) Single-gene qRT-PCR using mRNA harvested from shp53-HMEC left untreated or treated with either OSM alone or in combination with the small molecule TGFβR1 inhibitor, SB431542, and using primers targeting SNAI1, TNFSF10, MYC, and ATF3. (C) Single-gene qRT-PCR using primers targeting SNAI1 and mRNA from shp53-HMEC expressing shRNAs to GFP (shGFP), SMAD2 (shSMAD2), SMAD3 (shSMAD3), or SMAD4 (shSMAD4) grown in the presence (+) or absence (-) of recombinant OSM for 7 d.
![Figure 4. Persistent OSM/STAT3 signaling promotes SMAD target-gene transcription. (A) mRNA harvested from shp53-HMEC plated in the presence (+) and absence (-) of either recombinant OSM [10 ng/mL] or recombinant TGF-β1 [5 ng/mL] for 7 d was subjected to a targeted TGF-β Signaling-Targets qRT-PCR profiler array. Genes exhibiting a similar change in expression at least 2-fold following treatment with both OSM and TGF-β1 were selected and displayed to compare gene expression between untreated shp53-HMEC and shp53-HMEC treated with either OSM or TGF-β1. (B) Single-gene qRT-PCR using mRNA harvested from shp53-HMEC left untreated or treated with either OSM alone or in combination with the small molecule TGFβR1 inhibitor, SB431542, and using primers targeting SNAI1, TNFSF10, MYC, and ATF3. (C) Single-gene qRT-PCR using primers targeting SNAI1 and mRNA from shp53-HMEC expressing shRNAs to GFP (shGFP), SMAD2 (shSMAD2), SMAD3 (shSMAD3), or SMAD4 (shSMAD4) grown in the presence (+) or absence (-) of recombinant OSM for 7 d.](/cms/asset/bf011f87-dedb-4fe3-9758-6342a6020fe8/kccy_a_1259037_f0004_b.gif)
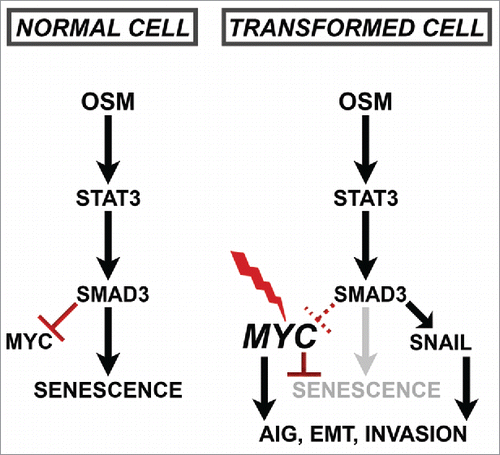
Figure 5. Constitutive MYC expression dismantles STAT3/SMAD3-induced senescence and cooperates with OSM to drive EMT. (A) Soft agar culture of shp53/MYC-HMEC plated in the presence or absence of recombinant OSM and recombinant TGF-β1 for 2 weeks. AIG was quantified by brightfield microscopy using Metamorph software. (B) Single-gene qRT-PCR analysis using primers targeting E-cadherin and Snail (encoded by CDH1 and SNAI1, respectively) and mRNA from shp53-HMEC and shp53/MYC-HMEC plated in the presence (+) or absence (-) of recombinant OSM and TGF-β1 for 7 d. (C) Western analysis of whole cell lysates from shp53/MYC-HMEC left untreated or treated with recombinant OSM for 7 d.
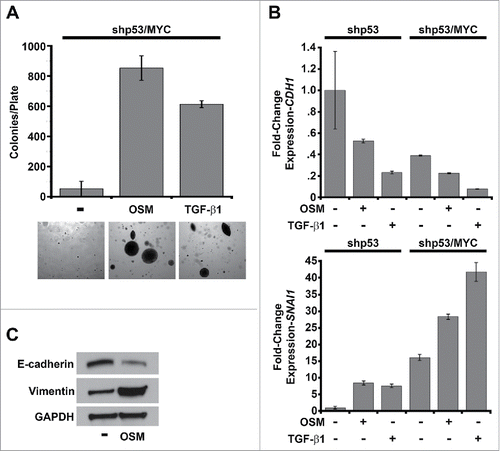
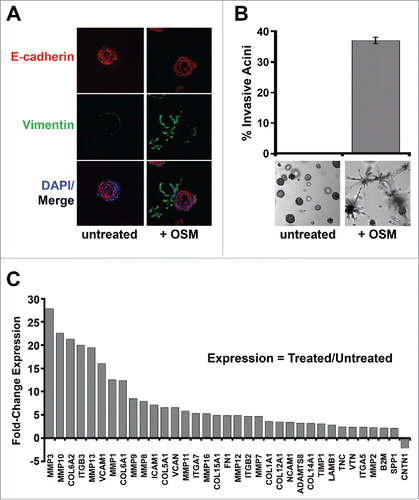
Figure 6. Constitutive MYC expression dismantles STAT3/SMAD3-induced senescence and cooperates with OSM to drive invasiveness. Shp53/MYC-HMEC were plated in 3D laminin-rich basement membrane (matrigel) culture in the presence and absence of recombinant OSM for 10 d. (A) Immunofluorescence using confocal microscopy at 100X magnification with Alexafluor-conjugated antibodies directed against E-cadherin (red) or Vimentin (green) and DAPI stain (blue, nuclear stain). (B) Invasive acini were photographed and quantified by brightfield microscopy at 5X magnification. (C) Following OSM-treatment, mRNA was harvested from shp53/MYC-HMEC after extraction from 3D matrigel culture and then subjected to a targeted Extracellular Matrix (ECM) and Adhesion Molecules qRT-PCR profiler array to compare gene expression differences between untreated and OSM-treated cells.