Figures & data
Table 1. Clinical features of the study population. Comparison s of normal (N, n = 20) and PE (PE, n = 24) pregnancies clinical features. Statistical significance (*) has been considered as p < 0.05.
Figure 1. p16INK4A and p18INK4C gene and protein expression in normal vs preeclamptic PDMSCs. (a) mRNA (left panel) and protein (right panel) expression of p16INK4A in normal (N, n = 20) and PE-PDMSCs (PE, n = 24) (b) mRNA (left panel) and protein (right panel) expression of p18INK4C in normal (N, n = 20) and PE-PDMSCs (PE, n = 24). Statistical significance (*) has been considered as p < 0.05.
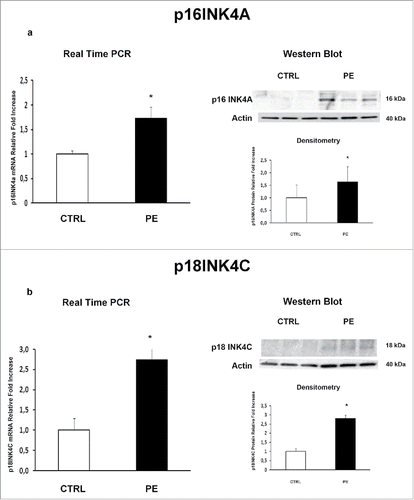
Figure 2. CDK4 and CDK6 gene and protein expression levels in normal vs preeclamptic PDMSCs. (A) mRNA (left panel) and protein (right panel) expression of CDK4 in normal (N, n = 20) and PE-PDMSCs (PE, n = 24) (B) (b) mRNA (left panel) and protein (right panel) expression of CDK6 in normal (N, n = 20) and PE-PDMSCs (PE, n = 24). Statistical significance (*) has been considered as p < 0.05.
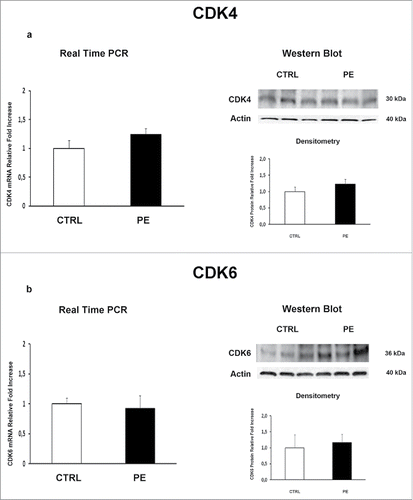
Figure 3. PDMSCs conditioned media effect on physiological term placental villous explants viability. Positive control (+) was provided by the kit. Explants treated by Triton X-100 and by unconditioned culture media for 8 h were used as high (H) and low controls respectively. Cytotoxicity in 72h supernatant of physiological villous explants treated with unconditioned media (CTRL) or media conditioned by normal (N CM) and preeclamptic (PE CM) PDMSCs was assessed by LDH assay. Statistical significance (*) has been considered as p < 0.05.
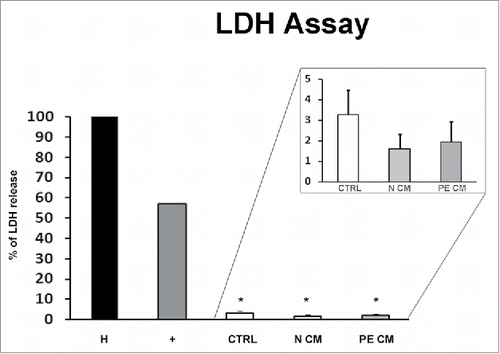
Figure 4. JunB and Cyclin D1 gene and protein expression levels in physiological placental villous explants treated with culture media conditioned by normal or PE-PDMSC. (A)JunB mRNA (left panels) and protein (right panels) expression levels in physiological villous explants treated with unconditioned media (CTRL, n = 16 explants) or media conditioned by normal (N CM, n = 16 explants) and preeclamptic (PE CM, n = 16 explants) PDMSCs as assessed by Real Time PCR and Western Blot analysis. B) Cyclin D1 mRNA (left panels) and protein (right panels) expression levels in physiological villous explants treated with unconditioned media (CTRL, n = 16 explants) or media conditioned by normal (N CM, n = 16 explants) and preeclamptic (PE CM, n = 16 explants) PDMSCs as assessed by Real Time PCR and Western Blot analysis. Statistical significance (*) has been considered as p < 0.05.
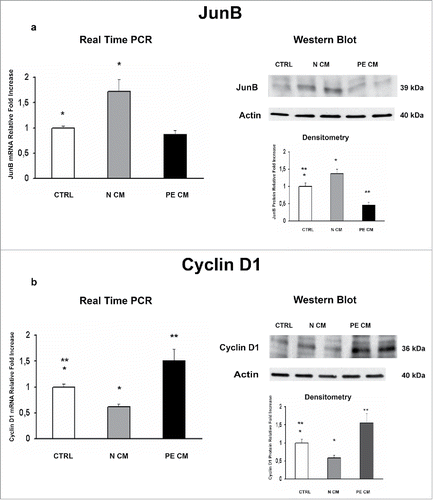
Figure 5. P16INK4A and p18INK4C gene and protein expression levels in physiological placental villous explants treated with culture media conditioned by normal or PE-PDMSC. (A) P16INK4A mRNA (left panels) and protein (right panels) expression levels in physiological villous explants treated with unconditioned media (CTRL, n = 16 explants) or media conditioned by normal (N CM, n = 16 explants) and preeclamptic (PE CM, n = 16 explants) PDMSCs as assessed by Real Time PCR and Western Blot analysis. B) and p18INK4C mRNA (left panels) and protein (right panels) expression levels in physiological villous explants treated with unconditioned media (CTRL, n = 16 explants) or media conditioned by normal (N CM, n = 16 explants) and preeclamptic (PE CM, n = 16 explants) PDMSCs as assessed by Real Time PCR and Western Blot analysis. Statistical significance (*) has been considered as p < 0.05.
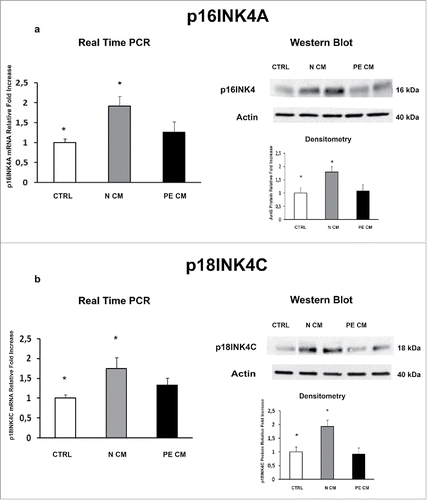
Figure 6. CDK4and CDK6 gene and protein expression levels in physiological placental villous explants treated with culture media conditioned by normal or PE-PDMSC. (A) CDK4 mRNA (left panels) and protein (right panels) expression levels in physiological villous explants treated with unconditioned media (CTRL, n = 16 explants) or media conditioned by normal (N CM, n = 16 explants) and preeclamptic (PE CM, n = 16 explants) PDMSCs as assessed by Real Time PCR and Western Blot analysis. B) CDK6 mRNA (left panels) and protein (right panels) expression levels in physiological villous explants treated with unconditioned media (CTRL, n = 16 explants) or media conditioned by normal (N CM, n = 16 explants) and preeclamptic (PE CM, n = 16 explants) PDMSCs as assessed by Real Time PCR and Western Blot analysis. Statistical significance (*) has been considered as p < 0.05.
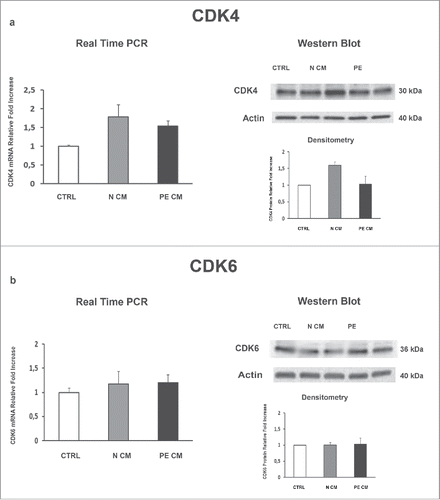
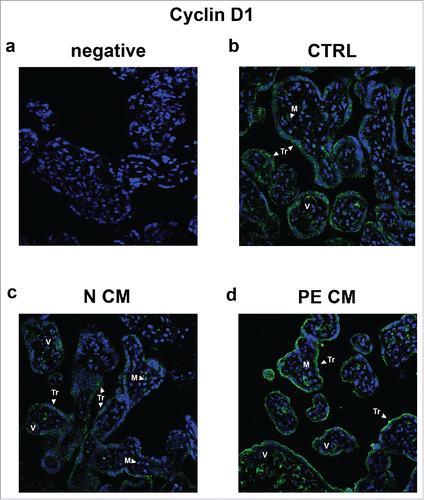
Figure 7. Cyclin D1 spatial localization in physiological placental villous explants treated with culture media conditioned by normal or PE-PDMSC. (A) Absence of positive immunoreactivity for Cyclin D1 in section stained with control IgG. B) Cyclin D1 spatial localization in physiological villous explants treated with unconditioned media (CTRL, n = 4 explants) assessed by immunofluorescent staining. C) Cyclin D1 spatial localization in physiological villous explants treated with media conditioned by normal (N CM, n = 4 explants) assessed by immunofluorescent staining. C) Cyclin D1 spatial localization in physiological villous explants treated media conditioned by with preeclamptic (PE CM, n = 4 explants) assessed by immunofluorescent staining. Cell nuclei are showed in blue by DAPI signal. TR, trophoblast cells; M, mesenchyme; V, vessel. Original magnifications, x40.