Figures & data
Figure 1. De novo lipogenesis in the cell. Abbreviations: MUSFA, monounsaturated fatty acids; PUSFA, polyunsaturated fatty acids; FADS, fatty acid desaturase; CE, cholesterol esters; TG, triglyceride; PL, phospholipid; TCA, tricarboxylic acid; ACC, acetyl-CoA carboxylase; ELOVL, elongation of very long chain fatty acids protein; FASN, fatty acid synthase; SCD1, stearoyl-CoA desaturase 1; ACLY, adenosine triphosphate citrate lyase; TCA, tricarboxylic acid. Details are reported in the main text.
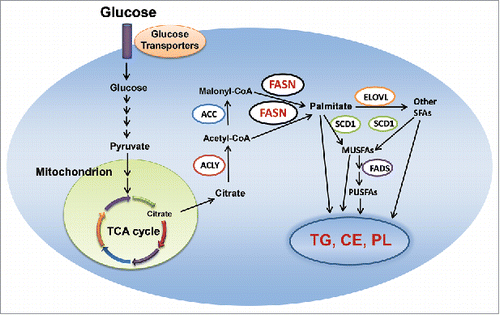
Figure 2. Regulation of de novo lipogenesis via the AKT/mTOR cascade. Abbreviations: RTKs, receptor tyrosine kinases; PTEN, phosphatase and tensin homolog; mTOR, mammalian target of rapamycin; mTOR, mTOR complex; SREBP-1, sterol regulatory element-binding protein-1. Details are reported in the main text.
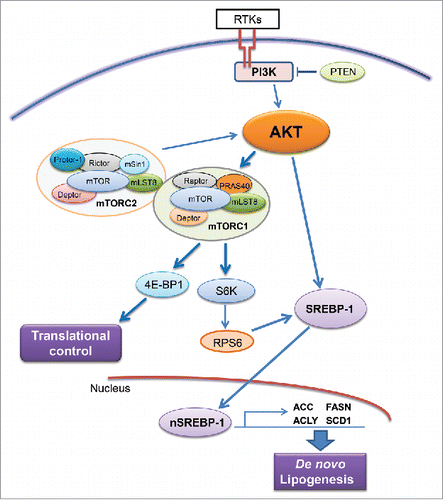
Figure 3. Levels of FASN mRNA along human hepatocarcinogenesis. (A) Expression levels of FASN in normal livers (NL; n = 12), surrounding non-tumorous liver tissues (SL; n = 133) and corresponding HCC samples (n = 133) as assessed by real-time RT-PCR. (B) Comparison of FASN levels in surrounding non-tumorous livers associated with different etiologic factors for HCC development. (C) Comparison of FASN levels in HCC associated with different etiologic factors involved in hepatocarcinogenesis. Tukey–Kramer's test: ***P<0.0001. Abbreviations: ASH, alcoholic steatohepatitis; HBV, hepatitis B virus; HCV, hepatitis C virus; NASH, non-alcoholic steatohepatitis.
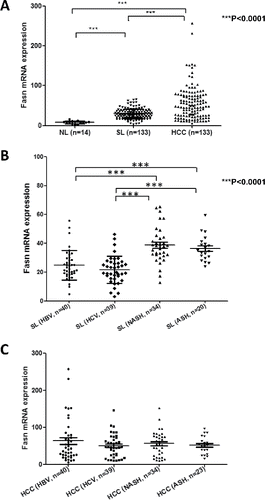
Figure 4. Expression of FASN and ACC is not elevated in c-Met/β-catenin mouse tumor tissues (right panel), when compared with HCCs induced by AKT/c-Met overexpression (middle panel). Scale bar: 200µm. Abbreviations: WT, wild-type; FASN, fatty acid synthase; ACC, acetyl-CoA carboxylase; HE, hematoxylin and eosin staining.
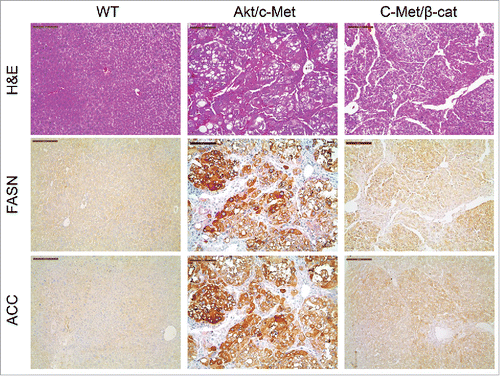
Figure 5. FASN expression is not upregulated along c-Met/β-catenin driven hepatocarcinogenesis. (A) Study design: activated forms of c-Met and β-catenin were hydrodynamically injected together with Cre into FASNfl/fl mice (c-Met/β-catenin/Cre, n = 4). This method allows the deletion of FASN while simultaneously expressing c-Met and β-catenin in the same FASNfl/fl hepatocytes. As a control, c-Met and β-catenin were injected in the same mice together with the empty vector (c-Met/β-catenin/pT3, n = 4). (B) Survival curve of c-Met/β-catenin/Cre mice (n = 4) and c-Met/β-catenin/pT3 injected FASNfl/fl mice (n = 4), P = 0.16. (C) Histologically, well to moderately differentiated HCC were found in both cohorts of mice and immunostaining demonstrated the low protein expression of FASN in c-Met/β-catenin/Cre HCC cells. Scale bar: 200µm for H&E and FASN, 100µm for Ki-67. Abbreviations: FASN, fatty acid synthase; HE, hematoxylin and eosin staining.
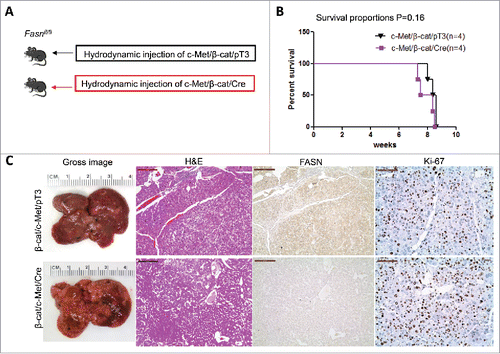
Figure 6. Genetic ablation of FASN in the mouse liver does not affect tumor development driven by c-Met/β-catenin co-expression. (A) Study design: Overexpression of c-Met/β-catenin promotes the development of multiple hepatocellular carcinomas both in FASNfl/fl mice with an intact FASN gene (n = 4) and in those where FASN was deleted by the Cre system (AlbCre;FASNfl/fl mice; n = 4). (B) c-Met/β-catenin induced HCC formation in wild-type and liver-specific FASN KO mice with the same latency and histology. (C) Survival curve of FASNfl/fl mice (n = 4) and liver-specific FASN KO mice (AlbCre;FASNfl/fl mice; n = 4) injected with c-Met/β-catenin, P = 0.35. Scale bar: 200µm for H&E and FASN, 100 µm for Ki-67. Abbreviations: FASN, fatty acid synthase; HE, hematoxylin and eosin staining.
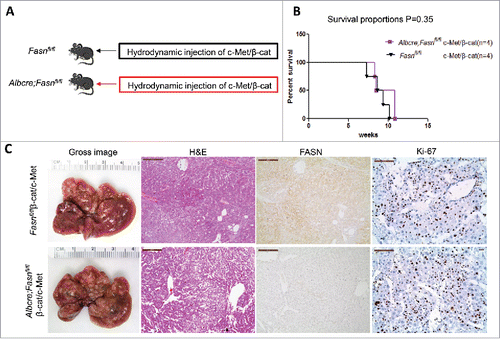
Table 1. Summary of the immunohistochemical patterns of FASN, ACC, and Ki-67 detected in wild-type livers (Wild-type), livers injected with empty vector (Empty vector) as well as in hepatocellular lesions (both preneoplastic and neoplastic) developed in AKT, AKT/c-Met, and c-Met/β-catenin mice.
