Figures & data
Figure 1. Unique binding motifs in the N-terminus of SLBP interact with FEM1A, FEM1B, and FEM1C. (A) Diagram representing the domain structure of SLBP. The amino acid sequence and substrate receptor binding motifs representing the “degron hotspot” are shown. TAD, translational activation domain; NLS, nuclear localization sequence; RBD, RNA binding domain. (B) FEM1A, FEM1B, and FEM1C interact with amino acids 1–99 of SLBP. C-E Mapping the FEM1A, FEM1B and FEM1C binding regions in SLBP. HEK293T cells were transfected with either empty vector (EV) or FS-tagged SLBP constructs. MLN4924 was added to the cells for 4 hours before collection. Cell lysates were affinity precipitated with anti-STREP resin, and affinity precipitations were probed with the indicated antibodies. (F) The ligase-deficient SLBP(ABCdegron) mutant is unable to bind to CTIF. HEK293T cells were transfected with FLAG-tagged SLBP constructs. Cell lysates were supplemented with SUPERase-In™ RNase Inhibitor and immunoprecipitated with anti-FLAG resin. The immunoprecipitations were probed with the indicated antibodies.
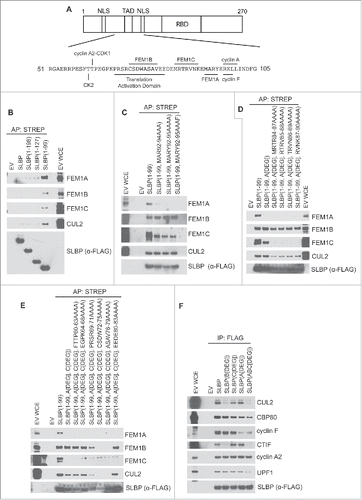
Figure 2. SLBP levels increase after depletion of FEM1A, FEM1B, or FEM1C. (A) Depletion of FEM1A, FEM1B, and FEM1C results in increased levels of SLBP in G1 phase. HeLa cells were synchronized at G1/S by double-thymidine block before trypsinization and release into fresh media. Cells were transfected with FEM1A, FEM1B, and FEM1C siRNA 48 hours before the first timepoint. Right: OligodT primed cDNAs corresponding to the immunoblot samples were analyzed by qPCR for FEM1A, FEM1B, and FEM1C mRNA. The data are presented as mean ± SD of one representative experiment. (B-D) Depletion of individual FEM1 proteins results in increased levels of SLBP in G1 phase. HeLa cells were synchronized at G1/S by double-thymidine block before trypsinization and release into fresh media. Cells were transfected with respective siRNAs 48 hours before the first timepoint. Samples were collected at the listed time points, and immunoblotted as indicated. *Asterisks denote non-specific bands.
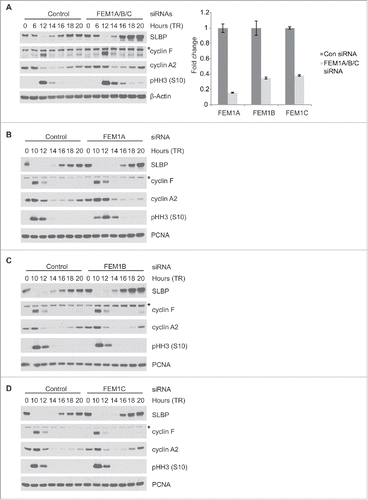
Figure 3. The interaction between SLBP and FEM1A, FEM1B, and FEM1C is required for SLBP degradation. (A) FLAG-SLBP(Bdegron), FLAG-SLBP(Cdegron), and FLAG-SLBP(Adegron) are each expressed at higher levels than FLAG-SLBP. HeLa cells infected with retroviruses expressing FLAG-tagged SLBP or FLAG-tagged SLBP mutants were synchronized at G1/(S)by double-thymidine block before trypsinization and release into fresh medium. Cells were collected at the indicated times, lysed, and immunoblotted. *Asterisk denote non-specific bands. (B) FLAG-SLBP(ABCdegron/Fdegron) is expressed at higher levels than FLAG-SLBP and FLAG-SLBP(Fdegron) and does not oscillate during the cell cycle. HeLa cells infected with retroviruses expressing FLAG-tagged SLBP, FLAG-tagged SLBP(Fdegron), or FLAG-SLBP(ABCdegron/Fdegron) were synchronized at the G1/(S)transition by double-thymidine block before trypsinization and release into fresh medium. Cells were collected at the indicated times, lysed, and immunoblotted.
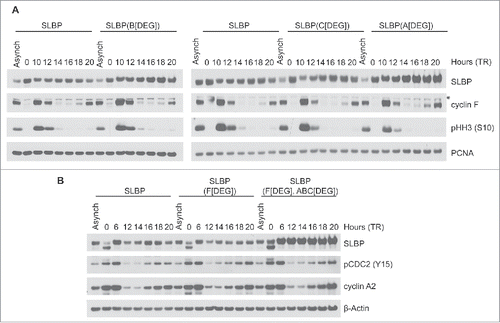
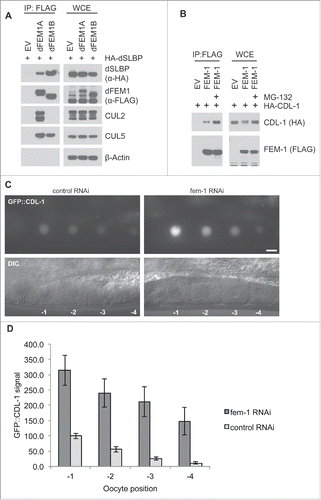
Figure 4. The FEM1-SLBP degradation pathway is conserved in metazoans. (A) FLAG-dFEM1A and FLAG-dFEM1B interact with HA-dSLBP. HEK293T cells were co-transfected with empty vector (EV), FLAG-tagged dFEM1A, or FLAG-tagged dFEM1B, and HA-tagged dSLBP. MLN4924 was added to the cells for 4 hours before collection. Cell lysates were immunoprecipitated with anti-FLAG resin, and immunoprecipitations were probed with the indicated antibodies. (B) FLAG-FEM1 interacts with HA-CDL-1. HEK293T cells were co-transfected with empty vector (EV) or FLAG-tagged FEM1. Where indicated, MG-132 was added to the cells for 4 hours before collection. Cell lysates were immunoprecipitated with anti-FLAG resin, and immunoprecipitations were probed with the indicated antibodies. (C) RNAi depletion of FEM-1 in C. elegans results in increased CDL-1 levels in oocytes. Representative epifluorescence images of GFP::CDL-1 expressed in the oocytes of hermaphrodite adults treated with fem-1 RNAi or control RNAi. The scale bar represents 10 uM. D. Graph showing the increase in GFP::CDL-1 levels in C. elegans oocytes after fem-1 RNAi treatment. The levels were normalized to the control RNAi signal in the oocyte at -1 position, which was set to 100. The graph represents mean ± SEM.