Figures & data
Figure 1. The expression of MALAT1 and HMGB1 in human OS cell lines. (A, B) The expression levels of MALAT1and HMGB1 in 5 OS cell lines, Saos2, MG63, U2OS, SW1353, HOS and normal cells, hFOB were examined by real-time qPCR assay. (C) A positive correlation between MALAT1 and HMGB1 was observed. (D) Two MALAT1 siRNAs, si-MALAT containing the interference site on 5274 and si-MALAT1 containing the interference site on 5074 were generated and transfected into OS cells to achieve MALAT1 knockdown. The inhibitory efficiency was verified using real-time PCR. (E) The expression levels of HMGB1 were monitored in response to 2 MALAT1 siRNAs using real-time PCR. Due to the higher inhibitory efficiency of Si-MALAT1 (5074), it was used for the further study. The data were presented as mean ± SD of 3 independent experiments.
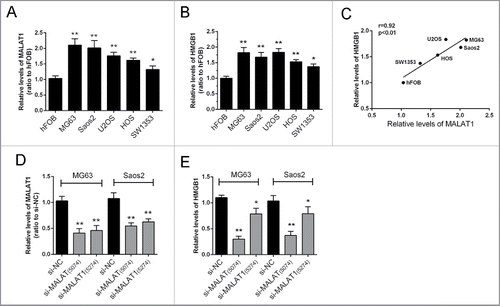
Figure 2. miR-142–3p/miR-129–5p correlated with MALAT1 and HMGB1. (A) By using TargetScan, miRanda and miRDB4 online prediction tools, candidate miRNAs that may bind to HMGB1 3′UTR were scanned out; among which 2 miRNAs were finally chosen for further experiments by using starBase. (B) The expression levels of miR-142–3p and miR-129–5p in Saos2 cells were determined in response to MALAT1 knockdown using real-time PCR. The data were presented as mean ± SD of 3 independent experiments.
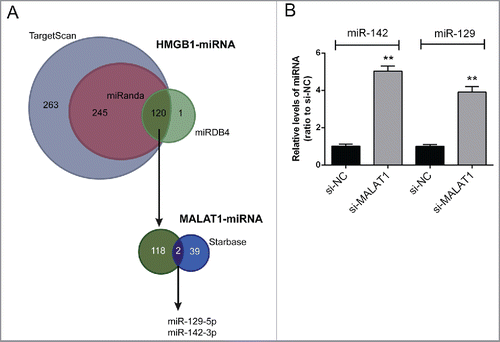
Figure 3. miR-142–3p/miR-129–5p correlated with MALAT1 through binding MALAT1. (A) miR-129–5p mimics or miR-129–5p inhibitor was transfected into Saos2 cells to achieve miR-129–5p overexpression or inhibition. The transfection efficiency was verified using real-time PCR. (B) miR-142–3p mimics or miR-142–3p inhibitor was transfected into OS cells to achieve miR-142–3p overexpression or inhibition. The transfection efficiency was verified using real-time PCR. (C) Two MALAT1 containing luciferase reporter constructs: wt-MALAT1 and a corresponding mut-MALAT1 which contains a 5 bp mutation in putative miR-142–3p and miR-129–5p binding site within MALAT1. These luciferase reporter constructs were co-transfected into Saos2 cells with miR-142–3p mimics/miR-129–5p mimics or miR-142–3p inhibitor/miR-129–5p inhibitor respectively. (D) and (E) The luciferase activity of the wt-MALAT1 reporter and mut-MALAT1 was monitored in different groups, compared with NC mimics or NC inhibitor group. The data were presented as mean ± SD of 3 independent experiments.
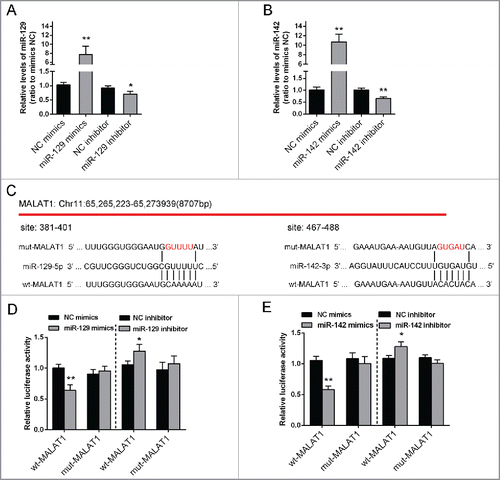
Figure 4. miR-142–3p/miR-129–5p regulated HMGB1 expression by binding its 3′ UTR. (A) and (B) The protein expression of HMGB1 was monitored in response to miR-129–5p or miR-142–3p overexpression or inhibition in Saos2 cells using Western blot assays. The data were presented as mean ± SD of 3 independent experiments. (C) and (E) Two HMGB1 containing luciferase reporter constructs: wt-HMGB1 and a corresponding mut-HMGB1 which contains a 5 bp mutation in one putative miR-142–3p and 2 miR-129–5p binding sites within its 3′-UTR. These luciferase reporter constructs were co-transfected into Saos2 cells with miR-142–3p mimics/miR-129–5p mimics or miR-142–3p inhibitor/miR-129–5p inhibitor respectively. (D) and (F) The luciferase activity of the wt-HMGB1 reporter and mut-HMGB1 was monitored in different groups, compared with NC mimics or NC inhibitor group. The data were presented as mean ± SD of 3 independent experiments.
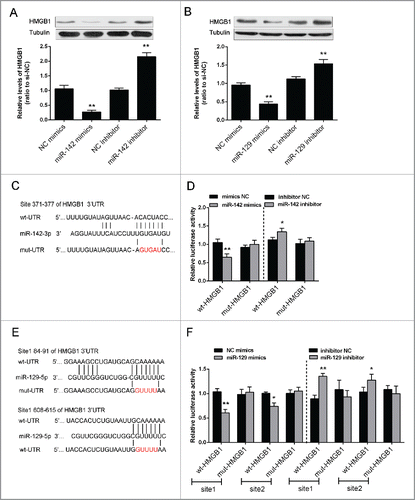
Figure 5. The role of MALAT1 and miR-142–3p/129–5p in OS proliferation and apoptosis (A) and (B) The protein expression of HMGB1 in MG63 and Saos2 was monitored in response to co-transfection with si-NC/si-MALAT1 and miR-142–3p/129–5p using Western blot assays. (C) and (D) The proliferation of MG63 and Saos2 cells was monitored in response to co-transfection with si-NC/si-MALAT1 and miR-142–3p/129–5p using MTT assays. (E) The apoptosis of MG63 and Saos2 cells was monitored in response to co-transfection with si-NC/si-MALAT1 and miR-142–3p/129–5p using Flow cytometer assays. The data were presented as mean ± SD of 3 independent experiments.
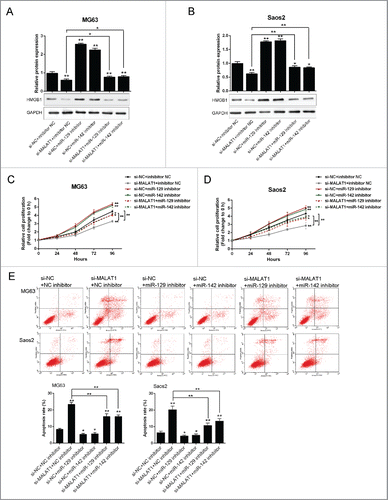
Figure 6. The expression differences of MALAT1/miR-142–3p/miR-129–5p/HMGB1 in tissues and the correlations. (A, B, C and D) 24 pairs of tumor and adjacent normal tissues were obtained, the expression levels of MALAT1, miR-142–3p, miR-129–5p and HMGB1 were determined. (D, E, F, G and I) An inverse correlation between the expression level of miR-142–3p and MALAT1, miR-142–3p and HMGB1, miR-129–5p and MALAT1, miR-129–5p and HMGB1, and a positive correlation between the expression level of MALAT1 and HMGB1 was observed. The data were presented as mean ± SD of 3 independent experiments.
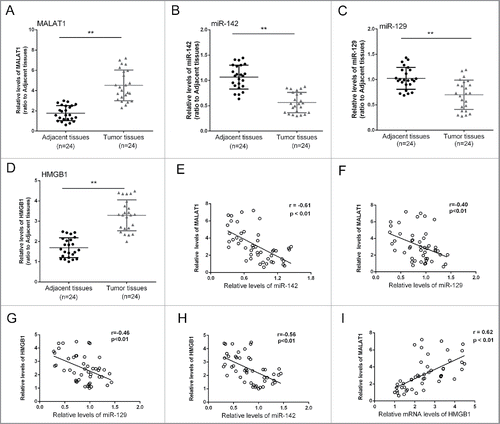
Table 1. The primers used for real-time PCR.
