Figures & data
Figure 1. Exploration of chemical derivatives of ONC201. (A) Structure of ONC201. (B) Core pharmacophore of ONC201 used for exploration of chemical derivatives. (C) ONC201 analogs including ONC identifier, R1 and R2 groups, and GI50 in HCT116 cells. N = 3; GI50 collected after 72 hours and calculated by PRISM. (D) Chemical structure and (E) ORTEP drawing of ONC212 crystals from X-ray data. X-ray crystallography was determined as described in methods.
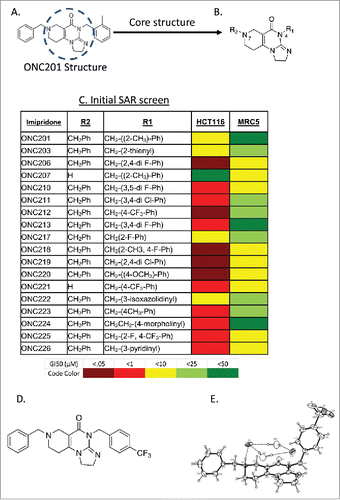
Figure 2. In vitro analysis of select analogs among several cell lines. A) Screen of GI50 values performed in lab of ONC212 and ONC206 compared to ONC201. B) GI50 in normal cell lines to determine toxicity in vitro. C) Colony assay of ONC201, ONC212, and ONC206 in select tissue types D) Representative colony assay in UACC-903. N = 3 for all experiments. ONC201: IC50 10 μM, IC25 2.5 μM; ONC212: IC50 0.01 μM, IC25 0.005 μM; ONC206: IC50 0.05 μM; IC25 0.01 μM) IC50 collected after 72 hours and calculated by PRISM. Colony assays where monitored until colonies formed.
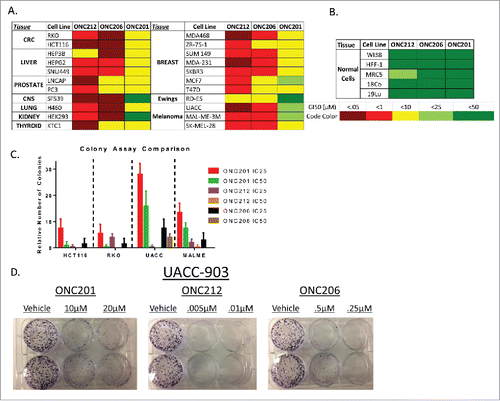
Figure 3. ONC212 and ONC206 downstream mechanism of action is similar to ONC201. A) CHOP and B) DR5 Gene expression data from ONC201, ONC206, and ONC212 treated in HCT116 cells over time relative to GAPDH and Vehicle. C) ONC201, ONC212, and ONC206 TRAIL surface expression analysis at indicated time points in HCT116 and TRAIL-resistant HCT116-Bax−/−. D) Western blot analysis results of HCT116 cells treated with ONC201 and ONC212 treated overtime. E) Quantitation of western blots (ONC201: 10 μM; ONC212: 0.01 μM; ONC206: 0.05 μM). N = 3; Western blot representative of 3 biological replicates.
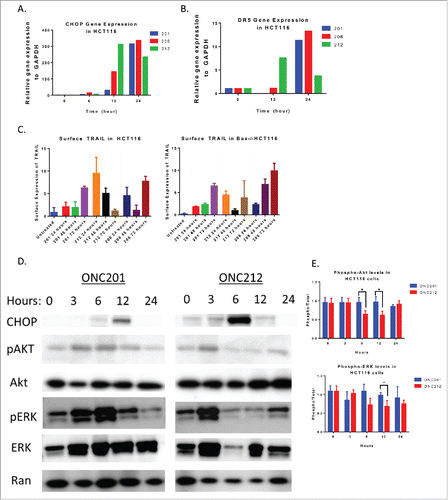
Figure 4. ONC212 and ONC06 induce cytotoxicity and inhibit migration and invasion similarly to ONC201. A) Representative cell cycle images in HCT116 cells. B) G1 and C) subG1 arrest in HCT116 cells at selected time points of ONC201 (10 μM), ONC212 (0.01 μM), and ONC206 (0.05 μM). D) Representative xCelligence migration assay. E) Cumulative Migration and F) Invasion using xCelligence software in select cell lines at GI50 values over 48 hours. ONC201: 10 μM; ONC212: 0.01 μM; ONC206: 0.05 μM. N = 3. Migration and cell cycle representative image of 3 biological replicates.
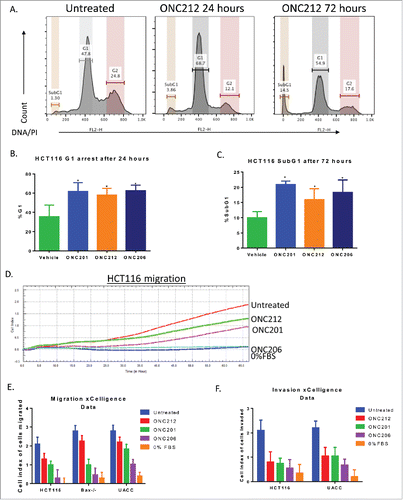
Figure 5. Pharmacokinetics and efficacy of oral ONC212. A) Pharmacokinetic profile through mass spectrometry at a dose of 125 mg/kg with blood extracted into a MeOH pellet through oral administration n = 3. Efficacy of ONC212 at 50 mg/kg in athymic nude mice. Comparison of IP vs oral in B) HT29 and C) HCT116 xenografts overtime D) Final tumor volume comparison of ONC212 in two CRC xenografts. HT29 after 30 days. HCT116 p53-null after 6 weeks. PK n = 3; efficacy n = 6. **p<.01
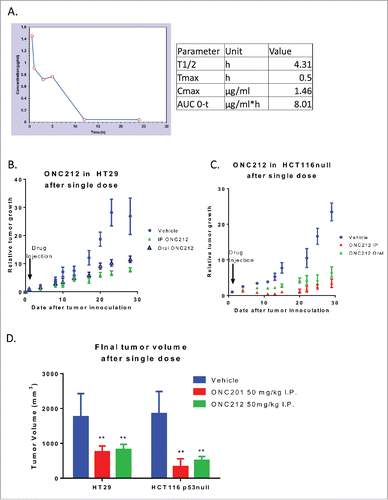
Figure 6. Genomic of Drug Sensitivity in Cancer (GDSC) Screen. A) In vitro sensitivity of 1068 human cancer cell lines to ONC212 (78nM – 20µM, 72h) organized by tumor type. The results are shown as ONC212 GI50 (nM) with representation of all cell lines in each tumor type. Dotted line represents average GI50 for each tumor type. The number of cell lines tested per tumor type are indicated. B) Average cell viability (area under curve, AUC) and C) IC50 with 72 hour ONC201 and ONC212 (0.078-20µM) treatment in a panel of 53 skin cancer cell lines in the GDSC screen. * indicates p < 6.68 × 10–24. D) Average cell viability (area under curve, AUC) and E) IC50 with 72 hour ONC212 (0.078-20µM) treatment in a panel of 15 wild-type (WT) and 32 BRAF V600E melanoma cell lines in the GDSC screen. P-value 0.0595
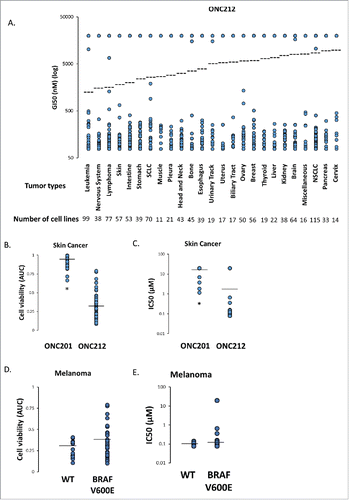
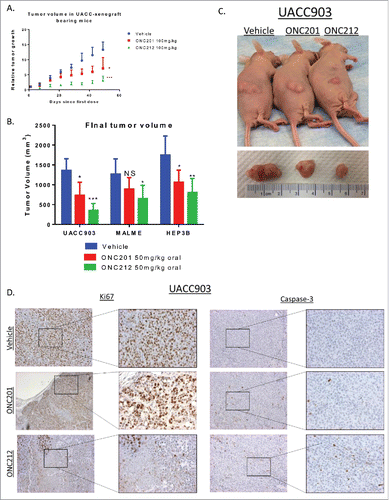
Figure 7. Efficacy of ONC212 in melanoma and liver cancer. A) relative tumor growth and B) Final tumor volume of MALME, UACC-903, and HEP3B xenograft bearing mice treated with Vehicle, ONC201, or ONC212 weekly. Final tumor volumes measured after 5 weeks for UACC903, 5 weeks for MALME, and 30 days for HEP3B as these where end of study. Statistics are compared to vehicle of each tissue type C) Photographic imaging of representative cohorts. D) Ki67 (left) and Caspase-3 (right) staining of selected cohorts at 5 weeks. N = 6 *p<.05; **p<.01 ***p<.005 IHC performed in 3 tumors from each cohort.