Figures & data
Figure 1. The structures of LCBs and ceramides. Shown are the structures of the LCBs, DL-dihydrosphingosine and phytosphingosine, and the ceramides, dihydroceramide and phytoceramide.
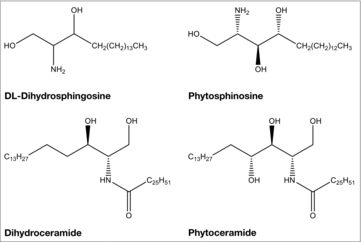
Table 1. Quantitative mating efficiencies.
Figure 2. VLCFA elongation is required for maintaining the appropriate sphingolipid composition in response to mating initiation. Radiolabeling of cells and subsequent separation of lipids by TLC determined the levels of various complex sphingolipids, inositolphosphorylceramides, and ceramides. Individual sphingolipid levels were assayed using scintillation counting and were compared with the total level of complex sphingolipid, and represented as a percentage. n = 5, *p < 0.01, **p < 0.001, ***p < 0.0001.
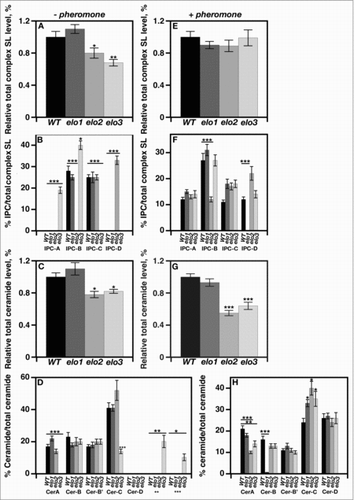
Figure 3. VLCFA elongation is required for shmoo formation in response to mating pheromone. 1 × 107 cells/ml were incubated with 20 μg/ml α factor and cell aliquots were collected at 0, 1, 2, 3, 4, hr. The percentages of shmoo formed per 300 cells were determined using light microscopy. bar1 cells (black circles); elo1 cells (open circles), elo2 cells (closed diamonds); elo3 cells (open diamonds). n = 5, *p < 0.01, ***p < 0.0001.
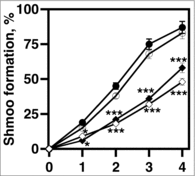
Figure 4. VLCFA elongation is required for cell cycle arrest during yeast mating. A-D, 1 × 107 cells/ml were incubated with 20 μg/ml α factor and cell aliquots were collected at 0, 1, 2, 3, 4, hr. Total RNA was used to determine the mRNA expression levels of CLN1 and CLN2 using qRT-PCR. CLN1 (white bars); CLN2 (black bars). E-H, cells were stimulated with 20 µg/mL α-factor. DNA content was measured using propidium iodide. Data was analyzed using BD CellQuest software. n = 5; G1 phase cells, black bars; S phase cells, light gray; G2M phase cells, dark gray.
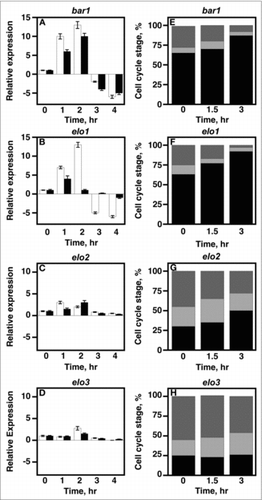
Figure 5. Bypassing the need for MAP kinase signaling does not suppress mating-specific gene expression defect of VLCFA elongation mutants. A FUS1 promoter-driven β-galactosidase reporter assay was used to examine transcriptional activity in the absence and presence of 20 μg/ml α-factor. Plasmid constructs containing the GAL1 promoter were used to overexpress Ste12, Ste4, Ste11ΔN, and Ste11-Cpr. Induction was performed using 2% galactose for 2 hr before addition of pheromone. n = 5; ***p < 0.0001.
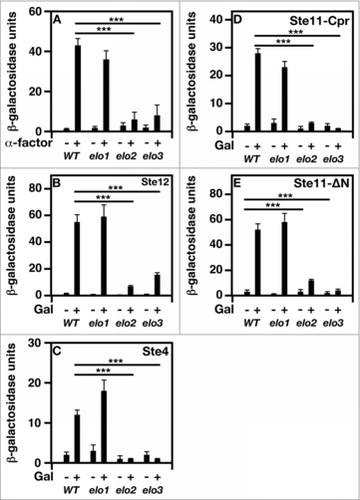
Figure 6. VLCFA elongation mutants lack MAP kinase-dependent activity. The levels of total and phosphorylated Fus3 were analyzed by western analysis using the appropriate antibodies. Actin level was used as a loading control. n = 5; pFus3, phosphorylated Fus3; Fus3-HA, total Fus3 protein.
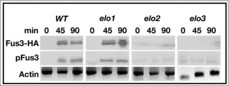
Figure 7. Loss of VLCFA elongation results in Ste5 mislocalization. (A-D), live cells were used to visualize GFP-Ste5. CEN URA3 STE5-GFPx3 was used to overexpress Ste5-GFPX3. Protein localization was visualized using a Leica DRME fluorescence microscope using 100X optics and a GFP filter. E, the percentage values of cellular localized Ste5-GFPX3 were based on examining 500 cells. n = 5; ***p < 0.0001.
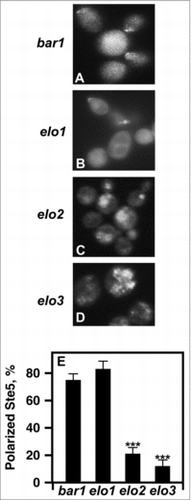
Figure 8. The expression of a plasma membrane-tethered Ste5 cannot re-activate mating-specific gene expression in VLCFA elongation mutants. A FUS1 promoter-driven β-galactosidase reporter assay was used to examine transcriptional activity in the absence and presence of 20 μg/ml α-factor and Ste5Q50L. n = 5; ***p < 0.0001.
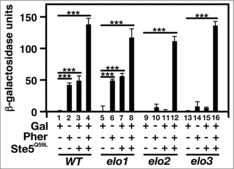
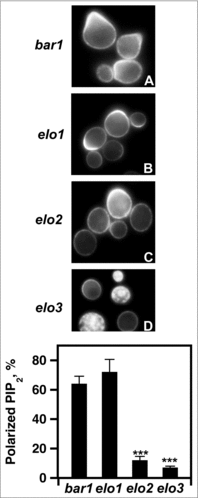
Figure 9. Loss of VLCFA elongation results in PIP2 mislocalization. (A-D), pGAL1-GST-GFP-PHPLCδ was used to express the PHPLCδ protein. For induction, cells were incubated with galactose for 2 hr. 20 μg/ml α factor was added 1 hr before protein visualization. PIP2 polarization was visualized using a Leica DRME fluorescence microscope using 100X optics and a GFP filter. E, the percentage values of properly localized PHPLC were determined using 500 cells. n = 5; ***p < 0.0001.