Figures & data
Table 1. Available threonine synthase (TS) structures. The table presents a list of all the structurally characterized TSs available at the PDB (release 3 May 2015), corresponding UniProt identifiers, TS class, zinc ribbon domain boundary*, and the organism name. *In case of Arabidopsis thaliana the residue numbers inside brackets “()” indicate the numbers in the PDB structure, while outside ones correspond to the UniProt sequence.
Figure 1. The domain boundaries in representative threonine synthase (TS) structures. Location of the lysine that binds PLP is marked in blue. Equivalent region in all TS joined by colored arrays. Abbreviations: Mt- Mycobacterium tuberculosis, At- Arabidopsis thaliana, Sc- Saccharomyces cerevisiae, Ec- Escherichia coli, Thr_syn_N- Threonine synthase N terminus (PF14821), ZnR- zinc ribbon, PALP- Pyridoxal-phosphate dependent enzyme catalytic domain (PF00291). In the case of A. thaliana, the residue numbers inside brackets “()” indicate the numbers in the PDB structure, while those outside correspond to the UniProt sequence.
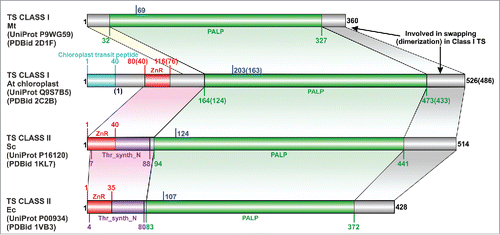
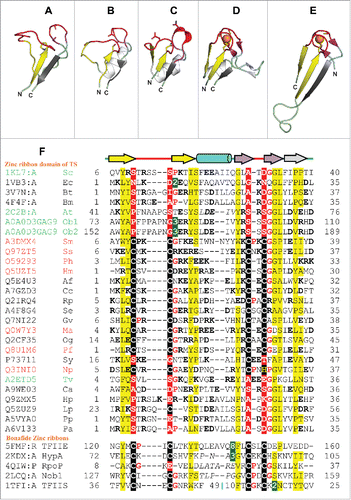
Figure 2. Structure and sequence comparison of TS zinc ribbons with bonafide zinc ribbon domains. (A-E) Ribbon diagrams of zinc ribbons from aTS (PDB identifier 2C2B_A), yTS (PDB identifier 1KL7_A), eTS (PDB identifier 1VB3_A), transcription initiation factor IIE α subunit (PDB identifier 1VD4_A), transcription factor IIS (PDB identifier 1TFI_A). In these figures, the N-terminal β-hairpin of the zinc ribbon is colored yellow, the C-terminal β-hairpin is purple, the additional β-strand that forms 3-stranded β-sheet with one of the hairpins is gray, and the zinc knuckles are red. The secondary structure elements (SSEs) that do not constitute the core of the zinc ribbon are colored white. Equivalent SSEs in (A-E) are colored alike. Zinc is shown as an orange sphere, and side chains of zinc-chelating aminoacids and equivalent residues in TS zinc ribbons are represented in stick form. (F) Structure-based multiple sequence alignment of representative TS zinc ribbons and other bonafide zinc ribbons. PDBid/UniProt identifier, organism name abbreviation, the first and the last residue numbers of the regions used in the alignment are indicated for each sequence. Diagrammatic representation of SSEs of the zinc ribbon is indicated above the alignment. Some sequence insertions are not shown, and the numbers of omitted residues are represented by numbers boxed in green. Potential metal-binding ligands are boxed in black and non-metal-binding residues at the same position are boxed in red. Uncharged aminoacids (all except Asp, Glu, Lys, and Arg) in mostly hydrophobic sites are highlighted yellow. Conserved small aminoacids (Pro, Gly) in the vicinity of zinc chelating residues are shown in red. UniProt/PDB identifier and organism name of sequences from eukaryotes, bacteria and archaea are shown in green, black and red, respectively. Sequence stretches where the structures are not superimposable have been italicized. The organism abbreviations are: Sc- Saccharomyces cerevisiae, Ec- Escherichia coli, Bt- Burkholderia thailandensis, Bm- Brucella melitensis, At- Arabidopsis thaliana, Ob- Oryza barthii, Sm- Staphylothermus marinus, Ss- Sulfolobus solfataricus, Ph- Pyrococcus horikoshii, Hm- Haloarcula marismortui, Af- Aliivibrio fischeri, Cc- Campylobacter curvus, Rp- Rhodopseudomonas palustris, Se- Saccharopolyspora erythraea, Gv- Gloeobacter violaceus, Ma- Methanocella arvoryzae, Og- Oceanicola granulosus, Pf- Pyrococcus furiosus, Sy- Synechocystis sp. PCC 6803, Np- Natronomonas pharaonis, Tv- Trichomonas vaginalis, Ca- Chloroflexus aurantiacus, Hp- Helicobacter pylori, Lp- Legionella pneumophila, Pp- Pseudomonas putida, Pa- Pseudomonas aeruginosa.