Figures & data
Figure 1. (A) A diagram of the pBI-SURF6 plasmid used for generation of stably transfected mouse NIH/3T3-174 fibroblasts capable to overexpress SURF6 in the presence of doxycycline (Dox). ampR – ampicillin-resistence gene, EGFP – the sequence coding for the EGFP protein, CMV – minimal CMV promoter, TRE – tetracycline-responsive element, SURF6 cDNA – cDNA of the mouse SURF6 gene, Surf6. (B) Immunoblots of control and induced NIH/3T3-174 fibroblasts after incubation without (-Dox) or with (+Dox) 100 ng/ml doxycycline for 24 and 48 hours. SURF6 was revealed with the anti-SURF6 serum,Citation26 β-tubulin (a loading control) was recognized with a commercial antibody. Star (*) indicates a position of a faster migrating SURF6 form appeared in +Dox fibroblasts. (C-J) General images of NIH/3T3-174 fibroblasts cultured without (-Dox; c, d) or with (+Dox; E-J) 100 ng/ml doxycycline for 48 (C-F and J) or 24 (G-I) hours. (C, E, and H) – EGFP fluorescence, (D and F) – phase contrast, (G and J) – immunolabeling for SURF6; (I) – DAPI staining of chromatin. In the absence of doxycycline, almost no EGFP fluorescence is seen (C). In the presence of doxycycline, the cells are intensely marked for EGFP (E and H), hereby indicating an effectiveness of the pBI-SURF6 plasmid. The phenotype of +Dox fibroblasts (F) remains similar to that of -Dox cells (D). SURF6 is distributed in nucleoli (G and J). A less intense anti-SURF6 immunolabeling of nucleoli is generally seen in cells with a weaker EGFP fluorescence (asterisks, G and H). Large-scale nuclear organization looks similar in different cells (I). Scale bars, (C-F) – 100 µm, (G-J) – 30 µm. (K) Trypan blue excision assay. Bar graphs illustrating the number of dead cells in –Dox (control, grey columns) and +Dox (black columns) cells after 24 and 48 hours of the induction. At each time-point, the control and experimental values do not differ statistically significant (p > 0.05). The data are shown as the mean ± SEM (n = 3).
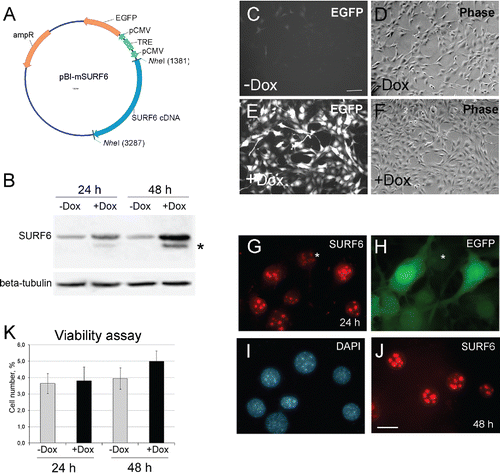
Figure 2. Growth rate of NIH/3T3-174 fibroblasts monitored in real time with the xCELLigence (RTCA)-DP instrument system. (A) Cells (∼3,000/well) were seeded in duplicate in an E-16 plate and cultured in the absence (-Dox, control) or the presence of 10 ng/ml, 100 ng/ml and 1000 ng/ml (+Dox) doxycycline. Doxycycline in DMEM (+Dox) or the equal volume of DMEM alone (-Dox) were added after 24 hours of cell seeding (arrow). The horizontal axis – time in hours from the beginning of the experiment. The vertical axis – the normalized cell index automatically registered every 15 min. Small vertical bars in the curves are fluctuations of the index at each registration time-point occurred in duplicates. Doxycycline in the concentration of 1000 ng/ml (pink curve) represses cell growth, while 10 ng/ml (blue curve) and 100 ng/ml (red curve) doxycycline stimulate proliferation as compared with -Dox cells (green curve). The SURF6 pro-proliferative effects start to be visible after 5–6 hours of the induction (corresponds to ∼30 hrs on the horizontal axis) and intensify with time. (B) Bar graphs illustrating the mean normalized cell index in +Dox cells in the percentage to control (-Dox) values considered as 100%. The proliferation rate was determined by analyzing the slopes of each curve obtained between the doxycycline administration (at ∼24 hrs of post-seeding) and the end of an experiment (∼70 hrs of post-seeding; n = 3). The results are expressed as the mean ± SEM. (*) indicate the values which differ insignificantly (p > 0.05). Induction of SURF6 overexpression with 10 ng/ml and 100 ng/ml doxycycline significantly (p < 0.05) activates proliferation as compared with non-stimulated NIH/3T3-174 fibroblasts, whereas 1000 ng/ml doxycycline turns out to be cytotoxic for the cells.
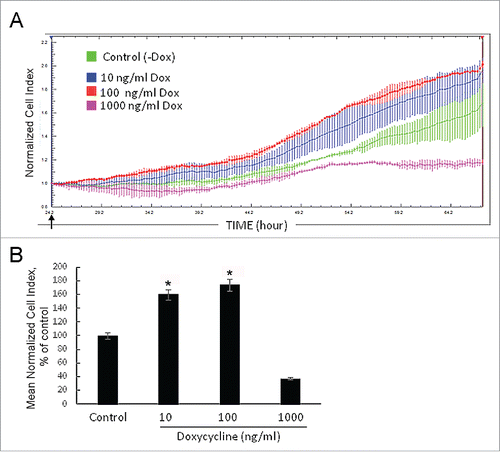
Figure 3. Effects of SURF6 overexpression on proliferation and viability of NIH/3T3-174 fibroblasts cultured without (-Dox) or with (+Dox) 100 ng/ml doxycycline up to 48 hours as analyzed with conventional phase contrast microscopy (A-F) and the MTT assay (G, H). (A-F) Cells were seeded in Petri dishes, cultured in complete growth medium for 3–4 hours to retrieve cell attachment to substrate. Twenty-five random fields of view were photographed shortly after doxycycline administration (“0” time-point, A) and 24 (B and C) and 48 (D and E) hours later, and representative images of the cells are shown in (A-E). Experiments were repeated five times, and in (F) bar graphs illustrating the mean cell number per field ± SEM are shown. The horizontal axis – time in hours, the vertical axis – the number of cells per square unit, small vertical bars – SEM. (G and H) Bar graphs illustrating MTT assay results. The horizontal axes – time in hours, the vertical axes in panel (G) – OD values at 570 nm, in panel (H) – the ODT /OD0 ratios equal to the OD values scored after 24 and 48 hours and normalized to the relative OD values at the “0” time-point. The data are presented as the mean ± SEM based on the results of three independent experiments.
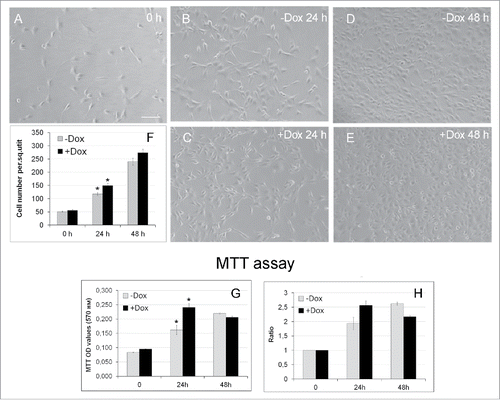
Table 1. Population doubling time (in hours) in different types of NIH/3T3 fibroblasts as calculated with the formula 1 (see Material and Methods for detail) after cell culturing without (-Dox) or with (+Dox) 100 ng/ml doxycycline for 24 or 48 hours. The data are expressed as the mean ± SEM (Standard Error of the Mean). At 24 hours of post-induction, the values observed in –Dox and +Dox NIH/3T3-174 fibroblasts differ statistically significantly (t-test, p < 0.001) and are shown in bold. The population doubling time in –Dox NIH/3T3-174, -Dox NIH/3T3-EGFP, +Dox NIH/3T3-EGFP and parental NIH/3T3 fibroblasts do not differ significantly (p > 0.05). At 48 hours, the population doubling-time between different fibroblasts differs insignificantly (p > 0.05).
Figure 4. Effects of SURF6 overexpression on cell cycle progression in NIH/3T3-174 fibroblasts as examined by flow cytometry after 24 hours of post-induction with 100 ng/ml doxycycline. (A and B) Results of a representative experiment, where cells were cultured without (-Dox, A) or with (+Dox, B) doxycycline. In the panel (C), bar graphs illustrate the mean percentage of +Dox (black columns) and -Dox (grey columns) cells which was calculated on the basis of four independent experiments. In G0/G1 phase the number of +Dox cells (46.2 ± 2.0%) is significantly lower (p < 0.05) that the number of -Dox cells (49.6 ± 1.9%), while in S phase the number of +Dox cells (44.9 ± 2.6%) significantly (p < 0.05) exceeds that of -Dox cells (40.5 ± 2.0%). The numbers of +Dox and –Dox cells at G2/mitosis are rather similar. Data are presented as the mean ± SEM.
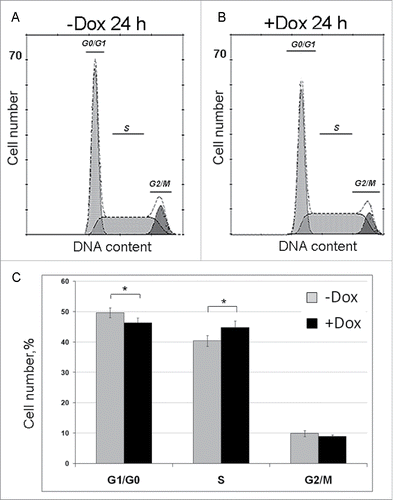
Table 2. Duration (in hours) of cell cycle phases in sub-confluent NIH/3T3-174 fibroblasts cultured without (-Dox) or with (+Dox) 100 ng/ml doxycycline for 24 hours after seeding. In brackets, the formulae used for calculations are indicated (see Materials and Methods for detail). The percentage of cells at G0/G1, S and G2/mitosis phases was determined by flow cytometry. The percentage of G0 cells was measured by cell population culturing with Br-dU for 24 hours. The number of G1 cells was as the number of G1/G0 cells minus the number of G0 cells. Data are expressed as the mean ± SEM. The compared pairs of values, which according to the t-test, differ significantly (p < 0.05) are indicated in bold. The duration of the cell division cycle in -Dox cells (17.6 ± 0.6) and in +Dox cells (14.0 ± 0.4) differ with p < 0.001.
Figure 5. The diagram of the primary 47S pre-rRNA in the mouse (A), Northern blot (B, C, and E) and qRT-PCR (F) analysis of rRNA expression in NIH/3T3-174 fibroblasts cultured without (-Dox) or with (+Dox) 100 ng/ml doxycycline for 24 hours. (A) 5′ETS, the 5′ external transcribed spacer; ITS1 and ITS2, the internal transcribed spacers 1 and 2 correspondingly; 18S, 5.8S and 28S are 18S, 5.8S and 28S rRNAs correspondingly. Small vertical bars and designations above the schemes indicate positions of the known pre-rRNA cleavage sites. Small horizontal bars above the schemes indicate positions of the PCR amplicons for 5′ETS and 18 S, 5.8 S, 28 S rRNA (according to ref. Citation1). (B and C) RNA was isolated from sub-confluent fibroblasts and hybridized with [P32] labeled probes recognizing the 18S rRNA (B) or ITS2 (C) regions of the 47S pre-rRNA transcript (A). Arrows indicate positions of the main rRNA species. (D) In mouse cells, processing of the 45S pre-rRNA proceeds mainly through two alternative pathways. In pathway 1, the pre-rRNA processing is initiated in the 5′-ETS to yield 41S pre-rRNA, while in pathway 2, the first cleavage occurs in ITS1 at site 2c to yield 34S rRNA (the longest precursor for 18S rRNA) and 36S rRNA (the longest precursor for 28S rRNA). The 41S pre-rRNA is further cleaved at site 2c into 20S and 36S rRNAs, which are then processed to 18S and 28S rRNAs correspondingly. (according to Citation1). (E) Bar graphs illustrating intensities of rRNA hybridization signals in +Dox cells (black columns) normalized to the values in -Dox cells (grey columns). Data in +Dox cells are presented as the mean ± SEM based on quantification of the results of four Northern blots. (F) Bar graphs illustrates the amount of the 47S pre-rRNA (the 5′ETS amplicons), 18S, 5.8S and 28S rRNAs in +Dox cells (black columns) normalized to the values in -Dox cells (grey columns). The values obtained in +Dox cells are presented as the mean N ± SEM based on the results of five independent experiments.
![Figure 5. The diagram of the primary 47S pre-rRNA in the mouse (A), Northern blot (B, C, and E) and qRT-PCR (F) analysis of rRNA expression in NIH/3T3-174 fibroblasts cultured without (-Dox) or with (+Dox) 100 ng/ml doxycycline for 24 hours. (A) 5′ETS, the 5′ external transcribed spacer; ITS1 and ITS2, the internal transcribed spacers 1 and 2 correspondingly; 18S, 5.8S and 28S are 18S, 5.8S and 28S rRNAs correspondingly. Small vertical bars and designations above the schemes indicate positions of the known pre-rRNA cleavage sites. Small horizontal bars above the schemes indicate positions of the PCR amplicons for 5′ETS and 18 S, 5.8 S, 28 S rRNA (according to ref. Citation1). (B and C) RNA was isolated from sub-confluent fibroblasts and hybridized with [P32] labeled probes recognizing the 18S rRNA (B) or ITS2 (C) regions of the 47S pre-rRNA transcript (A). Arrows indicate positions of the main rRNA species. (D) In mouse cells, processing of the 45S pre-rRNA proceeds mainly through two alternative pathways. In pathway 1, the pre-rRNA processing is initiated in the 5′-ETS to yield 41S pre-rRNA, while in pathway 2, the first cleavage occurs in ITS1 at site 2c to yield 34S rRNA (the longest precursor for 18S rRNA) and 36S rRNA (the longest precursor for 28S rRNA). The 41S pre-rRNA is further cleaved at site 2c into 20S and 36S rRNAs, which are then processed to 18S and 28S rRNAs correspondingly. (according to Citation1). (E) Bar graphs illustrating intensities of rRNA hybridization signals in +Dox cells (black columns) normalized to the values in -Dox cells (grey columns). Data in +Dox cells are presented as the mean ± SEM based on quantification of the results of four Northern blots. (F) Bar graphs illustrates the amount of the 47S pre-rRNA (the 5′ETS amplicons), 18S, 5.8S and 28S rRNAs in +Dox cells (black columns) normalized to the values in -Dox cells (grey columns). The values obtained in +Dox cells are presented as the mean N ± SEM based on the results of five independent experiments.](/cms/asset/9fce90ac-d56c-4786-ab06-df89fd18b0e2/kccy_a_1371880_f0005_b.gif)
Table 3. Primers and FAM-conjugated probes used in qRT-PCR analysis of the amplified DNA products (amplicons) corresponding to the mouse 47S pre-rRNA transcripts, 18S, 5.8S and 28S rRNAs, beta-actin and SURF6 mRNAs.
