Figures & data
Figure 1. Genome-wide two-hybrid screen for CENP-ACse4 identified Sgo1 as an interacting partner. (A) Schematic of yeast two-hybrid assay to identify interactors of CENP-ACse4 used here as a bait. BD = binding domain, AD = activating domain, UAS = upstream activating sequence. GAL4 gene, a transcription factor, produces BD and AD protein products, which are required for transcription of the reporter gene. GAL4-BD+CENP-ACse4 and GAL4-AD+Prey (yeast genes) fusion constructs were used. These fusion proteins alone cannot activate reporter gene transcription; however, expression of both fusion proteins allows interaction between CENP-ACse4 and prey protein leading to transcription of the reporter gene. (B) List of genes showing statistically significant two-hybrid interaction with CENP-ACse4.
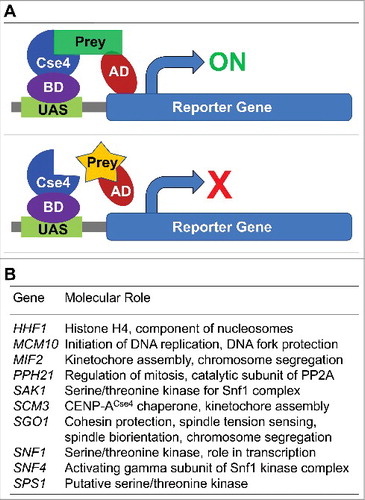
Figure 2. CENP-ACse4 interacts with Sgo1 and sgo1-NT in vivo and in vitro. (A) Sgo1 prey constructs was tested against the CENP-ACse4 bait construct. Sgo1 prey resulted in increased growth compared with vector (pADC) or CENP-ACse4 prey when tested against the CENP-ACse4 FL bait construct. (B) CENP-ACse4 interacts in vivo with Sgo1 and sgo1-NT. Wild type strain (SGO1-MYC, YMB10165), sgo1-NT-MYC (YMB10160) expressing CENP-ACse4 from galactose inducible promoter (GAL1) were grown in YEP with 2% galactose + 2% raffinose at 25°C. Untagged strain (OCF1533-4B), SGO1-MYC (AMY905), and sgo1-NT-MYC (YMB10076) were used as a control. Cell extracts were prepared for immunoprecipitation experiments using anti-HA agarose antibodies (A2095, Sigma-Aldrich). Eluted proteins were analyzed by Western blotting with anti-Myc (Sgo1-Myc or sgo1-NT-Myc; a-14, sc-789, Santa Cruz Biotechnology), anti-HA (CENP-ACse4; H6908, Sigma Aldrich), and anti-Tub2 (loading control) antibodies. IN = input, and IP = immunoprecipitated samples. (C) A minimal prey construct of AD-sgo1-NT132 (amino acids 1–132) resulted in two-hybrid positive interactions with CENP-ACse4-DBD, sgo1-NT (amino acids 1–150) and Rts1-DBD. Scm3-AD was also positive with CENP-ACse4-DBD. Two-hybrid selection plates were SD–HLT and supplemented with 3-aminotriazole (1 or 3 mM). (D) Affinity pull-down of recombinant proteins shows that GST-sgo1-NT132 immobilized on glutathione beads can interact and pull-down His6-CENP-ACse4 octasome from E. coli lysate. IN = input (1% of the input run on a separate gel). *Represents degraded or proteolyzed products. Pull-down experiments were performed three times with similar results.
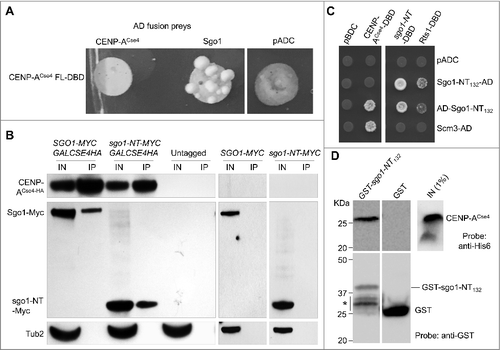
Figure 3. Sgo1 and sgo1-NT associate with core CEN in a cell cycle dependent manner. Wild type (SGO1-MYC, AMY905), sgo1-NT-MYC (YMB10076), and untagged control (OCF1533-4B) were grown in YPD to logarithmic phase (LOG) at 25°C, and synchronized in G1 with alpha factor (3 µM), S-phase with HU (0.2 M), and in G2/M with nocodazole (20 µg/mL) at 25°C for 2 hours. ChIP was performed using anti-Myc agarose beads (A7470, Sigma-Aldrich). Enrichment at CEN and at a negative control region ACT1 was determined by qPCR and is shown as % input. Average from three biological replicates ± standard error is shown. **p value <0.01, Student's t test. (A) Western blotting showing expression of Sgo1-Myc and sgo1-NT-Myc in LOG or various stages of the cell cycle. Antibodies used were: anti-Myc (Sgo1 or sgo1-NT; a-14, sc-789, Santa Cruz Biotechnology), and anti-Tub2 (loading control) antibodies. (B) FACS profiles show DNA content representing various stages of the cell cycle. (C) ChIP-qPCR showing enrichment levels of Sgo1-Myc and sgo1-NT-Myc at CEN1 in LOG phase or at various stages of the cell cycle. (D) ChIP-qPCR showing enrichment levels of Sgo1-Myc and sgo1-NT-Myc at CEN3 in LOG phase or at various stages of the cell cycle. (E) ChIP-qPCR showing enrichment levels of Sgo1-Myc and sgo1-NT-Myc at CEN5 in LOG phase or at various stages of the cell cycle. (F) ChIP-qPCR showing enrichment levels of Sgo1-Myc and sgo1-NT-Myc at a negative control region ACT1 in LOG phase or at various stages of the cell cycle.
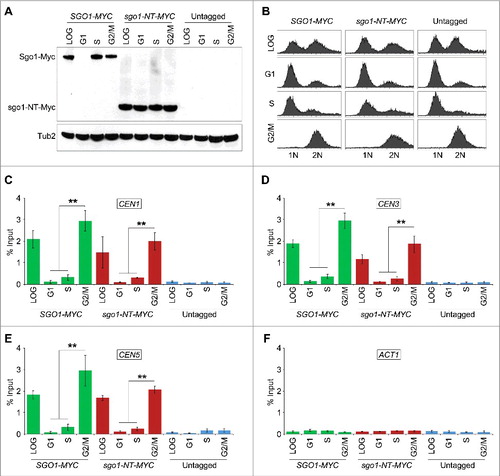
Figure 4. sgo1-NT associates with core CEN but not with peri-CEN chromatin. (A) Schematic of CEN and peri-CEN regions on chromosome III. Centromere (CEN3, violet oblong), cohesin associated regions (CARs; green triangle with vertical lines) are shown. CEN is located at 114 kb; peri-CEN regions 115 (1 kb from CEN), 112 (2 kb from CEN), 134 (20 kb from CEN), and chromosome arm CAR 261 (147 kb from CEN) were examined. (B) Association of Sgo1-Myc and sgo1-NT-Myc at CEN and peri-CEN regions. ChIP samples from cells synchronized in G2/M as described in were used. Enrichment at CEN3 and peri-CEN regions was determined by qPCR and is shown as % input. Average from three biological replicates ± standard error is shown.
Figure 5. Association of Sgo1 and sgo1-NT with core CEN requires CENP-ACse4. Wild type strain (SGO1-MYC, YMB10165), and sgo1-NT-MYC (YMB10160) expressing HA-tagged CENP-ACse4 from galactose inducible promoter (GAL1) were grown in YEP with 2% galactose + 2% raffinose at 25°C (Cse4-ON). Untagged strain (OCF1544-4B) was used as a control. Cells were collected, washed in dH2O and grown at 25°C in YEP with 2% glucose for 3 hours to Shut-off the expression of CENP-ACse4 (Cse4-OFF). Samples were collected for DNA content, protein extraction, and ChIP analyses. (A) CENP-ACse4 protein levels are not detectable upon its depletion. Western blot analysis was carried out on whole cell protein extracts prepared from cultures grown under Cse4-ON and Cse4-OFF conditions as described above. Blots were probed with anti-HA (CENP-ACse4; clone 12CA5, Roche Molecular Systems), and anti-Myc (Sgo1-Myc or sgo1-NT-Myc; a-14, sc-789, Santa Cruz Biotechnology) antibodies. (B) Depletion of CENP-ACse4 causes synchronization of cells in G2/M phase of the cell cycle as revealed by FACS analysis. (C) CEN levels of CENP-ACse4 are reduced upon its depletion. ChIP for CENP-ACse4 was carried out using anti-HA agarose beads (A2095, Sigma-Aldrich). Enrichment at CEN1, CEN3, CEN5, and ACT1 (negative control) was determined by qPCR and is shown as % input. Average from three biological replicates ± standard error is shown. **p value <0.01, *p value <0.05, Student's t test. (D) Sgo1 and sgo1-NT fail to remain associated with CEN chromatin upon depletion of CENP-ACse4. ChIP for Sgo1-Myc and sgo1-NT-Myc was performed using anti-Myc agarose beads (A7470, Sigma-Aldrich). Enrichment at CEN1, CEN3, CEN5, and ACT1 (negative control) was determined by qPCR and is shown as % input. Average from three biological replicates ± standard error is shown. **p value <0.01, *p value <0.05, Student's t test.
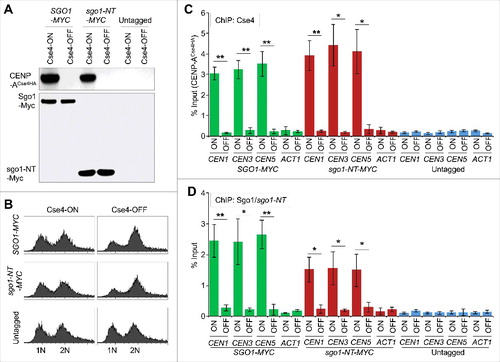
Figure 6. The N-terminus of Sgo1 (sgo1-NT) is required for faithful chromosome segregation. (A) Schematic of full-length SGO1 and its mutant alleles. sgo1-NT includes amino acids residues 1–150 (deletion of amino acids 151–591), whereas, sgo1-CT includes amino acids residues 151–591 (deletion of amino acids 2–150). Symbols: CC denotes the N-terminus coiled coil domain (amino acids 43–87); B denotes the C-terminus basic SGO1 motif (amino acids 366–390), and pink vertical line represents destruction box (amino acids 494–498). (B) sgo1-NT is required for faithful chromosome segregation independently of C-terminus basic domain. Frequency of CF loss in wild type (SGO1; YPH1018), sgo1Δ (YMB10224), sgo1-CT (YMB10221), and sgo1-NT (YMB10225) strains was determined as described in Materials and Methods. At least 1000 colonies from three independent transformants were counted. Average from three biological experiments ± standard error. Values sharing the same letter are not significantly different at a 5% level based on the analysis of variance (p > 0.05).
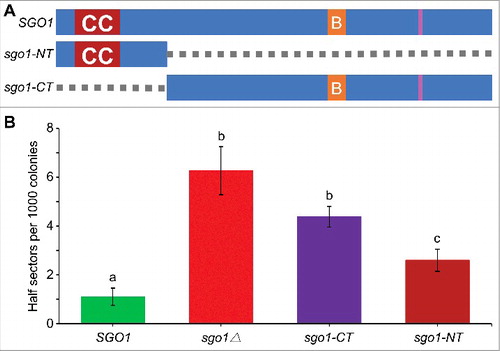
Table 1. List of yeast strains, plasmids, and primers used in this study.
