Figures & data
Figure 1. shRNAs derived from 17 TS genes cause growth reduction in HeyA8 cells.
(A) Percent cell confluence over time of HeyA8 cells after infection with shScr, shL3 and two shRNAs for each of the 17 TS genes. The curves for cells infected with two independent shRNAs for each TS gene and their specific ID number and respective growth reduction caused by each shRNA are shown in blue and green. Percent growth reduction values (as shown in ) were calculated using STATA1C software when cells infected with shScr reached half maximal confluency as indicated by the red dotted line. (B) Percent cell confluence over time of HeyA8 cells after infection with shScr, shL3, and five shRNAs targeting PTEN (left) and four shRNAs targeting p53 (right). Percent growth reduction values are shown in .
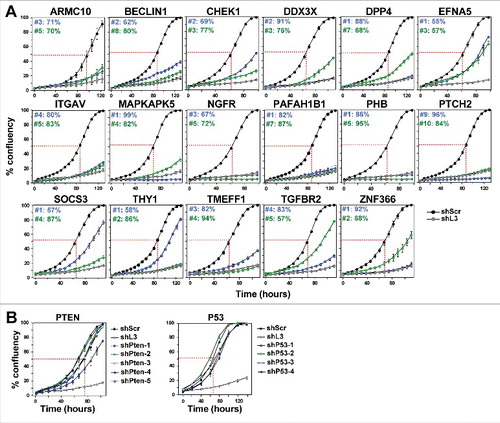
Table 1. Summary of all assays leading to the identification of DISE inducing TS-derived shRNAs
Figure 2. Toxic shRNAs derived from eight TS genes induce DISE-like morphological changes in HeyA8 cells.
Representative phase-contrast images showing elongated cell shapes (A); enlarged, flattened cells and presence of intracellular granules in HeyA8 cells infected with shRNAs against eight of the 17 TS and shL3 (B). shScr treated cells are shown as control.
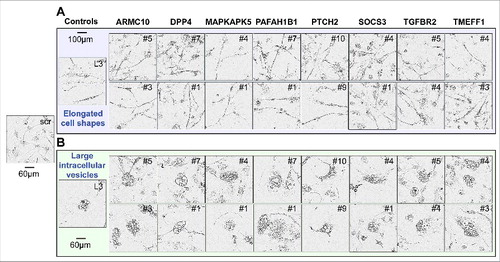
Figure 3. Toxic shRNAs derived from seven TS genes induce death that is biochemically similar to DISE.
Quantification of ROS production by dichlorofluorescin (DCFH) fluorescence (A), caspase-2 activity (B) and quantification of cell death with PI staining (C) in HeyA8 cells 8 days after infection with shScr, shL3, and shRNAs derived from eight of the TS genes. ID numbers are shown in . p-values were calculated using students t-test. *p < 0.01, **p < 0.001, ***p < 0.0001. For genes in black, both shRNAs were functionally active whereas genes shown in grey, only one of the two shRNAs had a significant effect. Color code for the two shRNAs per gene is the same as in .
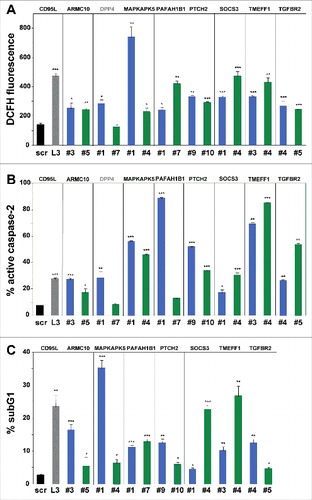
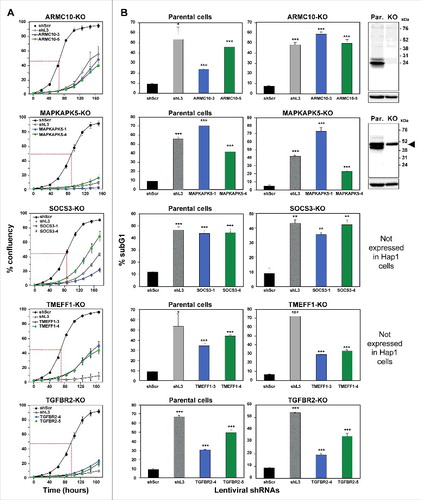
Figure 4. shRNAs derived from four TS genes kill cells in the absence of the transcript and/or protein produced from the targeted gene.
Percent cell confluence over time (A) and percent nuclear PI staining (B) of HAP1 parental cells, or HAP1 knock-out cells, after infection with shScr, shL3, or one of two shRNAs each targeting the respective TS. Percent growth reduction values (as shown in ) were calculated using STATA1C software when cells infected with shScr reached half maximal confluency as indicated by the red dotted line. p-values were calculated using a t-test. Western blot analyses in (B) confirm the knock-out of ARMC10 and MAPKAPK5 at the protein level. Arrowhead marks likely unspecific band. *p < 0.01, **p < 0.001, ***p < 0.0001. Color code for the two shRNAs per gene is the same as in .