Figures & data
Table 1. Clinical features of study population.
Figure 1. LOC100507600 is down-regulated in HSCR. (A) The expression of LOC100507600 in HSCR tissues (n = 64) and control tissues (n = 64). LOC100507600 was significantly reduced in patient tissues compared with control tissues. (B): Receiver Operating Characteristic (ROC) curve for the LOC100507600 to distinguish HSCR cases from controls.
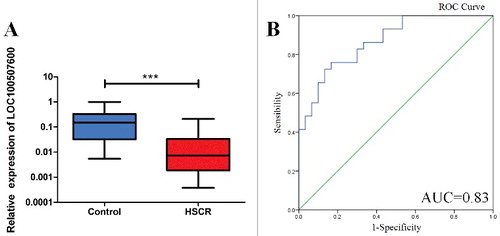
Figure 2. Cytobiology change after treating cells with LOC100507600 siRNA. (A) Human 293T and SH-SY5Y cell lines were transfected with LOC100507600 siRNA and then qRT-PCR is repeated three times to determine the efficiency of transfection. (B) Human 293T and SH-SY5Y cell lines were transfected with LOC100507600 siRNA to regulate its expression levels and cell proliferation was detected using the CCK8 assay. Knockdown of LOC100507600 suppressed cell proliferation. (C) Transwell assay was performed as described in method and indicated that down-regulation of LOC100507600 delayed cell migration. Pictures were captured under a light microscope with the magnification, × 10. (D and E)Flow cytometry demonstrated that the down-regulation of LOC100507600 had no effect on cell cycle progression and apoptosis.
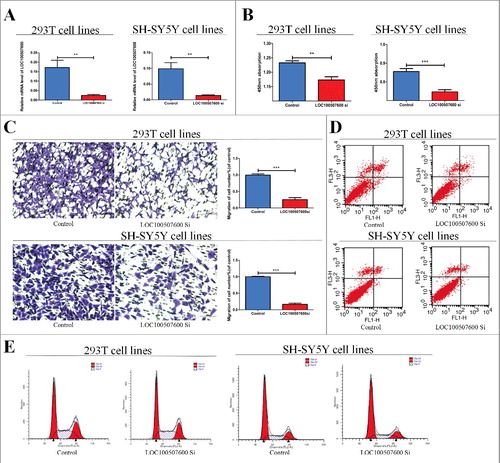
Figure 3. LOC100507600 serves as a sponge for miR128–1-3p. (A) The levels of nuclear control transcript (U6), cytoplasmic control transcript (GAPDH), and LOC100507600 were assessed by qRT-PCR in nuclear and cytoplasmic fractions. (B) The expression of miR128–1-3p in HSCR tissues and normal tissues. miR128–1-3p was significantly rose in patient tissues compared with normal tissues. (C) Superstratum:sequence alignment of human miR128–1-3p with LOC100507600. Bottom: mutations in the LOC100507600 sequence to create the mutant luciferase reporter constructs. (D) Luciferase reporter assay in 293T and SH-SY5Y cells after transfected with negative control or miR128–1-3p mimics, renilla luciferase vector pRL-SV40 and the reporter constructs. Both frefly and renilla luciferase activities are measured in the same sample. Firefly luciferase signals were normalized with renilla luciferase signals.
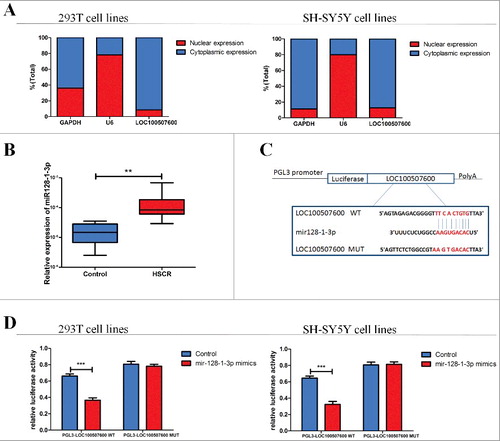
Figure 4. LOC100507600 regulates the miR-128–1-3p target, BMI1. (A) Relative expression of BMI1 in HSCR tissues in comparison with control tissues. BMI1 was significantly reduced in patient tissues. (B) Protein level of BMI1 in HSCR tissues and normal control samples were detected by Western Blot assay. (C) Bivariate correlation analysis of the relationship between LOC100507600 and BMI1 expression level. (D) Superstratum:the putative miRNA binding sites in the BMI1 sequence. The putative miRNA recognition sites were cloned downstream of the luciferase gene and named pGL3-BMI1-Wild. Bottom: mutations in the BMI1 sequence to create the mutant luciferase reporter constructs named pGL3-BMI1-Mut. (E) Left: The luciferase reporter in 293T cell lines. Right: the luciferase reporter in SH-SY5Y cells. Luciferase activity was determined using the dual luciferase assay and shown as the relative luciferase activity normalized to renilla activity.
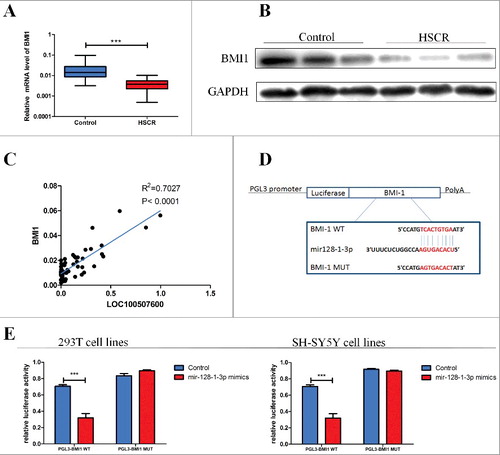
Figure 5. LOC100507600-miR128–1-3p regulatory loop is critical for cell function. (A) LOC100507600 siRNA with or without miR128–1-3p inhibitor was transfected into 293T cells and the relative expression of BMI1 was evaluated by qRT-PCR, × 20 (B) Western blot analysis of BMI1 protein level following treatment of 293T and SH-SY5Y cells with LOC100507600 siRNA or with LOC100507600 siRNA plus miR128–1-3p inhibitor. GAPDH was used as control. (C and D) CCK8 assay and Transwell assays were performed to determine the proliferation and migration of miR128–1-3p transfected cells and treated with LOC100507600 siRNA plus miR128–3p inhibitor.
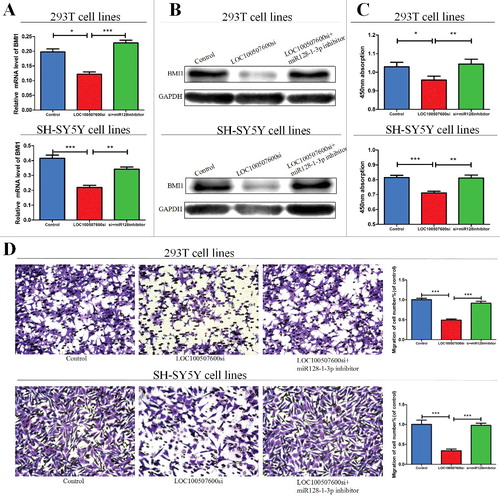
Table 2. Sequences of primers for qRT-PCR and siRNA related sequence.
