Figures & data
Figure 1. LncRNA ZFAS1 was upregulated in CRC tissues and associated with poor prognosis. (A,B) Relative expression of ZFAS1 measured by qRT-PCR in CRC tissues and adjacent non-tumor tissues. (C) Kaplan-Meier analysis and log-rank test were performed to assess the prognosis of CRC patients with high- and low- ZFAS1 expression. (D) ROC curve showed the AUC value of ZFAS1 (0.837, 95% CI: 0.713-0.921). *p<0.05
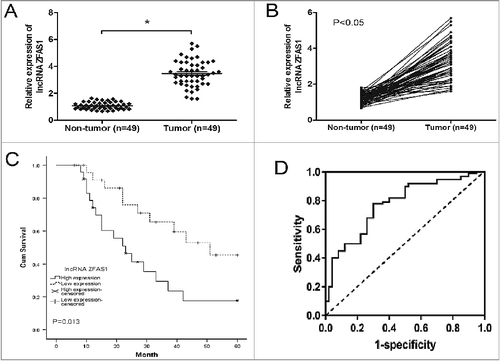
Table 1. Correlation between ZFAS1 expression and clinicopathological features of CRC.
Figure 2. LncRNA ZFAS1 inhibition suppressed CRC cells proliferation and invasion. (A) Relative expression of ZFAS1 measured by qRT-PCR in CRC cells and human colonic epithelial cells HCoEpiC. (B) ZFAS1 expression was examined by qRT-PCR assay in cells transfected with si-ZFAS1 or si-NC. (C) CCK-8 assay showed that ZFAS1 depletion inhibited the proliferation of SW480 and HT-29 cells. (D) colony formation assay revealed that ZFAS1 inhibition suppressed the clone formation of SW480 and HT-29 cells. (E) Transwell invasion assay suggested that ZFAS1 suppression inhibited the invasion ability of SW480 and HT-29 cells. *p<0.05
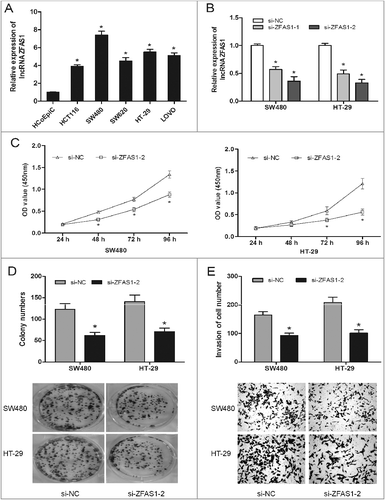
Figure 3. LncRNA ZFAS1 knockdown suppressed tumor growth in vivo. (A) SW480 cells transfected with sh-ZFAS1 were subcutaneously injected into back of nude mice. Tumor volume was measured every week. (B,C) The subcutaneous tumors were taken photos (B) and weighted (C) in sh-ZFAS1 group or sh-NC group. *p<0.05
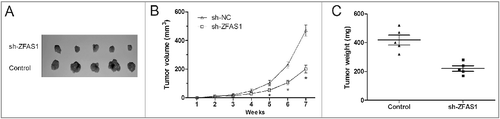
Figure 4. LncRNA ZFAS1 directly interacted with miR-507 in CRC. (A) The predicted complementary sequence between miR-484 and ZFAS1 3’-UTR. (B) HEK293T cells were transfected with ZFAS1 3′-UTR Wt or Mut reporter plasmids along with miR-484 mimics, and then luciferase activity assays were performed. (C) Relative expression of miR-484 was measured by qRT-PCR in CRC tissues. (D) Relative expression of miR-484 was measured by qRT-PCR in CRC cells. (E) Pearson's correlation analysis between miR-484 and ZFAS1 expression in CRC tissues. (F) miR-484 expression in ZFAS1 knockdown CRC cells was determined by qRT-PCR. (G) Association of ZFAS1 and miR-484 with Ago2 in CRC cells. *p<0.05
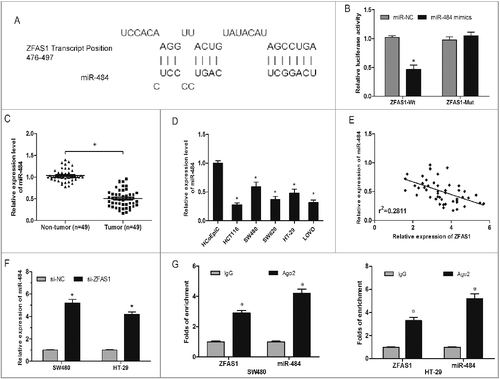
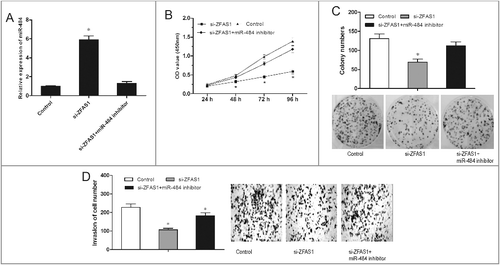
Figure 5. miR-484 reversed the effect of ZFAS1 on CRC tumorigenicity. (A) Relative expressions of miR-484 in CRC cells transfected with si-ZFAS1 and miR-484 inhibitor. (B) CCK-8 assay showed that proliferation of CRC cells transfected with si-ZFAS1, si-ZFAS1+miR-484 inhibitor. (C) Colony formation assay showed the clone number of CRC cells transfected with si-ZFAS1, si-ZFAS1+miR-484 inhibitor. (D) Transwell invasion assay showed the invasion of CRC cells transfected with si-ZFAS1, si-ZFAS1+miR-484 inhibitor. *p<0.05