Figures & data
Figure 1. UQCRFS1 pathway from synthesis of the precursor in the cytoplasm, transport into the mitochondrial matrix through the TOM/TIM23 import system, incorporation of the [2Fe-2S] clusters in the matrix and translocation to pre-cIII2 in the inner membrane (IM). According to our model, the N-terminal MTS is cleaved off in situ by the UQCRC1+UQCRC2 MPP activity. See main text for more details.
![Figure 1. UQCRFS1 pathway from synthesis of the precursor in the cytoplasm, transport into the mitochondrial matrix through the TOM/TIM23 import system, incorporation of the [2Fe-2S] clusters in the matrix and translocation to pre-cIII2 in the inner membrane (IM). According to our model, the N-terminal MTS is cleaved off in situ by the UQCRC1+UQCRC2 MPP activity. See main text for more details.](/cms/asset/f43f0858-c864-48d6-b4b4-f313f7d8b3e9/kccy_a_1417707_f0001_c.jpg)
Figure 2. Graphic alignment of the Rieske protein precursors from yeast (S. cerevisiae), fruit fly (D. melanogaster), chicken (G. gallus) and mammals (bovine, mouse and human). Numbers indicate relevant residue positions. The two yeast cleavage sites, at positions 22 and 30 are indicated with arrows. The main cleavage site, conserved in all organisms is indicated with the big arrow. Different color MTS indicates the lack of homology of these sequences, while the homology in the mature protein sequences in very high.
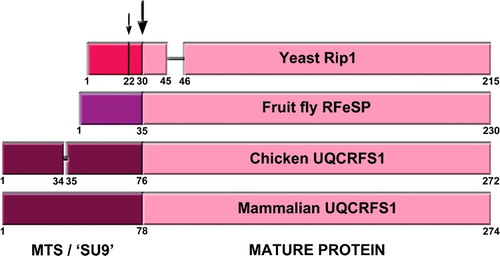
Figure 3. Structures of full complex III dimer (cIII2) and of isolated UQCRFS1 (of only one of the monomers), from bovine (PDB ID: 1BGY) and chicken (PDB ID: 3H1J). The arrows indicate the presence of the UQCRFS1 fragments in the cavity between UQCRC1 and UQCRC2. Images were generated with MacPyMol.
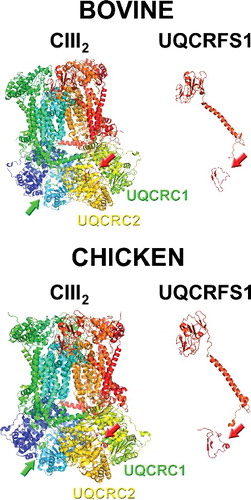
Table 1. UQCRFS1 MTS sequences found in the published cIII2 structures
