Figures & data
Figure 1. Exogenous RCC1 evicts Sgo1 and AurB from KTs in mitotic XEEs. (A) Experimental timeline. XEEs containing DSN (10,000 units/µl) were induced for DNA replication before driven back to mitosis. XEEs aliquots were analyzed by immunoblotting (IB) and immunofluorescence (IF) after indicated treatments. (B) Input reactions and chromatin fraction from untreated XEEs or XEEs with increasing concentrations of RCC1 (2, 6, or 20 μg/ml) were analyzed by IB analysis. Cycling XEEs were prepared and chromatin was isolated from different time points. Proteins content from both total extracts and chromatin fractions were analyzed by IB. The chromatin bound RCC1 intensity readings were quantified, normalized to first lane, and plotted in bottom panel. (C)(D) Chromatin from samples as in (B) were purified, stained with Hoechst 33342, and subjected to IF analysis against Bub1 (top, red) and (C) Sgo1 or (D) AurB (middle, green). Scale bar, 20 μm.
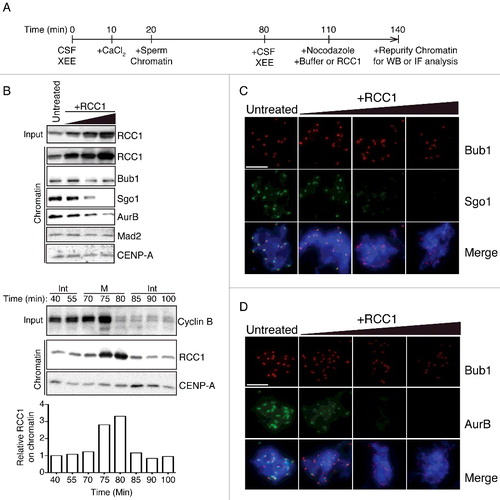
Figure 2. RCC1 evicts Sgo1 and AurB without blocking their recruitment. (A) Experimental timeline. XEEs aliquots were taken before (T0) and after (T30) RCC1 addition (20 μg/ml) and were analyzed by IB and IF analysis. (B) Chromatin fraction from XEEs were isolated at T0 and T30 and analyzed by IB analysis. (C)(D) Chromatin from samples as in (B) were purified, stained with Hoechst 33342, and subjected to IF analysis against Bub1 (top, red) and (C) Sgo1 or AurB (D) Histone H2ApThr121 (middle, green). In parallel, CSF XEEs depleted of Bub1 were also cycled to mitosis and subjected to the same IF analysis. Scale bar, 20 μm.
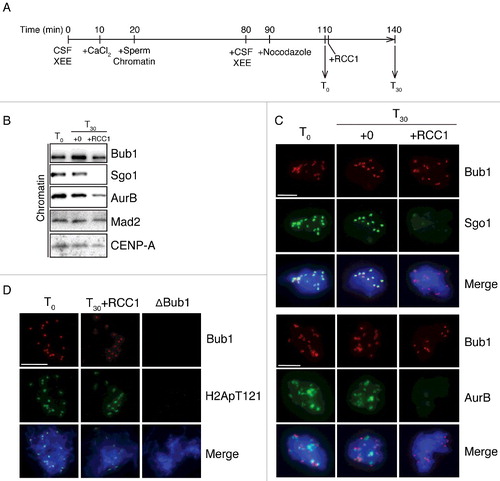
Figure 3. Eviction of inner KT proteins by RCC1 is independent of RanGEF activity. (A) Buffer, RanQ69L (0.2 mg/ml), RCC1WT (40 μg/ml), or RCC1Ran (40 μg/ml) were added to mitotic XEEs containing chromatin (10,000 DSN/μl). Input reactions and chromatin fractions from each sample were examined by IB analysis. (B) Chromatin from samples as in (A) were purified, stained with Hoechst 33342, and subjected to IF analysis against Bub1 (row 2, red) and Sgo1 (row 3, green). Scale bar, 20 µm. (C) Either buffer or nocodazole (20 μg/ml) were added to CSF-XEEs, as indicated. DSN were added to a concentration of 10,000 units/μl. Buffer, RanQ69L (20 μM), RCC1WT (40 μg/ml), or RCC1Ran (40 μg/ml) were added as indicated. CaCl2 was added to each sample to a concentration of 0.1 mg/ml. XEE aliquots before and after CaCl2 treatment were examined by IB.
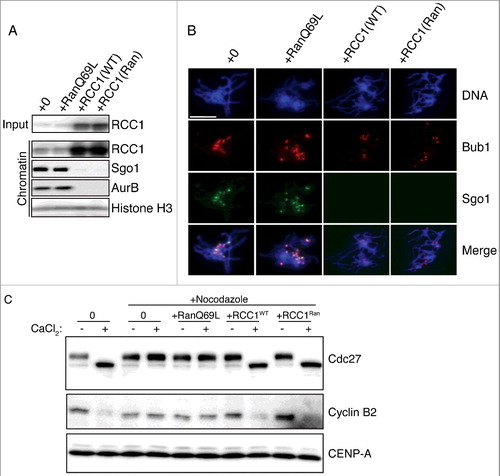
Figure 4. Mutant analysis shows that RCC1 must bind chromatin for eviction of inner KT proteins. (A) Table showing names, mutations, chromatin affinity (measured in XEEs), and RanGEF activity (measured through in vitro assays) of wild type or mutant RCC1. (B) 1 µM recombinant Ran charged with [α-32P]GTP was incubated with 0, 0.1, 0.3, or 1 nM RCC1 at RT for 30 min, and exchange was monitored using a filter retention assay. Radioactivity retained on filters was measured by scintillation counter and plotted against RCC1 concentration as mean ± SEM (n = 3). (C) Analysis of chromatin binding for RCC1 mutants. RCC1 was immunodepleted from XEEs. DSN were added to a concentration of 10,000 units/μl. Wild type or mutated RCC1 was added back to depleted XEEs at concentrations of 6, 20, 60, 200, or 600 μg/ml. Chromatin was re-purified and subjected to quantitative IB with antibodies against RCC1. (D) Fluorescent intensity of RCC1 in chromatin fractions from samples as in (C) were acquired by Odyssey infrared imaging system and plotted against input RCC1 concentration as mean ± SEM (n = 3). (E) Buffer, RCC1WT (40 µg/ml), RCC1Hist (40 µg/ml) or RCC1DNA (40 µg/ml) were added to mitotic XEEs containing chromatin (10,000 DSN/µl). Input reactions and chromatin fractions from each sample were examined by IB analysis.
![Figure 4. Mutant analysis shows that RCC1 must bind chromatin for eviction of inner KT proteins. (A) Table showing names, mutations, chromatin affinity (measured in XEEs), and RanGEF activity (measured through in vitro assays) of wild type or mutant RCC1. (B) 1 µM recombinant Ran charged with [α-32P]GTP was incubated with 0, 0.1, 0.3, or 1 nM RCC1 at RT for 30 min, and exchange was monitored using a filter retention assay. Radioactivity retained on filters was measured by scintillation counter and plotted against RCC1 concentration as mean ± SEM (n = 3). (C) Analysis of chromatin binding for RCC1 mutants. RCC1 was immunodepleted from XEEs. DSN were added to a concentration of 10,000 units/μl. Wild type or mutated RCC1 was added back to depleted XEEs at concentrations of 6, 20, 60, 200, or 600 μg/ml. Chromatin was re-purified and subjected to quantitative IB with antibodies against RCC1. (D) Fluorescent intensity of RCC1 in chromatin fractions from samples as in (C) were acquired by Odyssey infrared imaging system and plotted against input RCC1 concentration as mean ± SEM (n = 3). (E) Buffer, RCC1WT (40 µg/ml), RCC1Hist (40 µg/ml) or RCC1DNA (40 µg/ml) were added to mitotic XEEs containing chromatin (10,000 DSN/µl). Input reactions and chromatin fractions from each sample were examined by IB analysis.](/cms/asset/c79888d9-d350-4eca-a37b-0bc218864fee/kccy_a_1442630_f0004_oc.jpg)
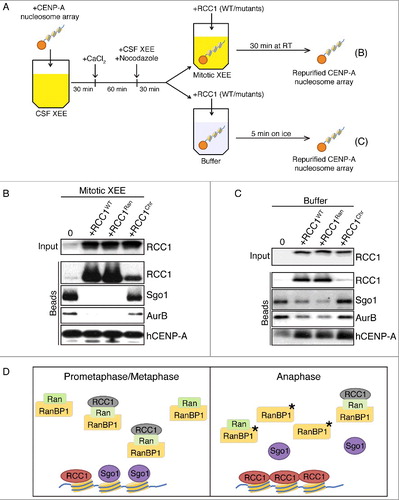
Figure 5. RCC1 evicts proteins from KTs assembled on CENP-A nucleosome arrays. (A) Schematic diagram with timeline. Streptavidin beads conjugated with CENP-A-containing nucleosome array were added into CSF-XEEs and cycled to mitosis. The beads were either maintained in M-phase XEEs (B) or isolated in buffer (C). Wild type or mutated RCC1 proteins were added, and the beads were isolated. (B) Buffer, RCC1WT (40 µg/ml), RCC1Ran (40 µg/ml) or RCC1Hist (40 µg/ml) were added to 100 µl mitotic XEEs containing 10 µl CENP-A-containing nucleosome array beads, and incubated at RT for 30 min. Beads were isolated, eluted, and examined by IB. (C) 10 µl CENP-A-containing nucleosome array beads were isolated from mitotic XEE and resuspended in 100 µl ice-cold washing buffer. Buffer, RCC1WT (40 µg/ml), RCC1Ran (40 µg/ml), or RCC1Hist (40 µg/ml) were added and incubated on ice for 5 min. Beads were isolated again, eluted, and examined by IB. (D) Model for the role of RCC1 in Sgo1 and CPC eviction: Prometaphase/metaphase: RCC1 is partitioned between an active, chromatin-bound pool (green) and an inactive pool (gray). The inactive pool is associated with RRR complexes that also contain RanBP1 and nucleotide-free Ran (green). Anaphase: phosphorylation of RanBP1 (asterisk) releases RCC1 from the RRR complex. The free RCC1 is recruited to chromatin and evicts inner KT proteins, including Sgo1, by physical competition.