Figures & data
Figure 1. (A) Meiotic DSB formation. The SPO11-TOPVIBL complex induces DSBs at genomic locations enriched for H3K4me3 and H3K36me3, histone modifications deposited by PRDM9. DSBs activate the ATM kinase, which suppresses SPO11 from further DSB formation by an as yet unknown mechanism. In response to DSBs, ATM phosphorylates histone H2AX (γ-H2AX) in surrounding chromatin. (B) Meiotic DSB processing and detection. SPO11, covalently attached to DNA, is removed by endonucleolytic cleavage by the MRE11 complex. This reaction releases SPO11 bound to short oligonucleotides (SPO11-oligo complexes) and exposes short single-stranded DNA (ssDNA) tails at DSB sites. These tails are further extended by resection and become substrates for DMC1 and RAD51 strand exchange proteins, which mediate homologous recombination. SPO11 oligos, ssDNA, and DMC1/RAD51-coated ssDNA can be analyzed by deep sequencing to map genome-wide DSB positions (see main text for references)
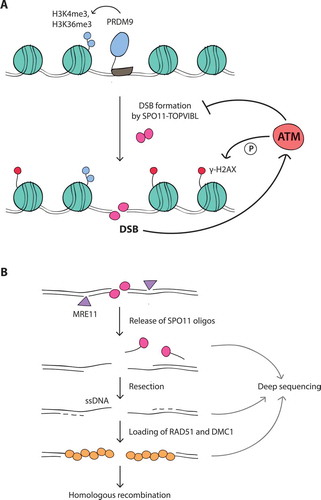
Figure 2. (A) Local DSB patterning. (top) Examples of SPO11-DSB hotspots on chromosome 1 in Atm wild type (wt) and Atm null (data from ref [Citation17].). Note the emergence of a new hotspot in the absence of ATM. (bottom) Overlays of Atm wt and Atm null hotspot profiles show a wider distribution of SPO11 oligos around the center of hotspot B in Atm null. Methods: As previously described [Citation17], hotspots were defined as regions in smoothed SPO11-oligo maps with uniquely mapped reads exceeding 50 times the genome average. The hotspot center was defined as the coordinate of the maximum SPO11-oligo value within the hotspot. Here, SPO11-oligo maps were smoothed with a 101-bp Hann filter. RPM denotes reads per million.(B) Schematic of MRE11-independent formation of SPO11 oligos by introduction of nearby DSBs (see main text)
![Figure 2. (A) Local DSB patterning. (top) Examples of SPO11-DSB hotspots on chromosome 1 in Atm wild type (wt) and Atm null (data from ref [Citation17].). Note the emergence of a new hotspot in the absence of ATM. (bottom) Overlays of Atm wt and Atm null hotspot profiles show a wider distribution of SPO11 oligos around the center of hotspot B in Atm null. Methods: As previously described [Citation17], hotspots were defined as regions in smoothed SPO11-oligo maps with uniquely mapped reads exceeding 50 times the genome average. The hotspot center was defined as the coordinate of the maximum SPO11-oligo value within the hotspot. Here, SPO11-oligo maps were smoothed with a 101-bp Hann filter. RPM denotes reads per million.(B) Schematic of MRE11-independent formation of SPO11 oligos by introduction of nearby DSBs (see main text)](/cms/asset/65592ade-52ec-4fe5-8241-da9ffbdfb1f5/kccy_a_1464847_f0002_oc.jpg)
Figure 3. Large-scale DSB patterning
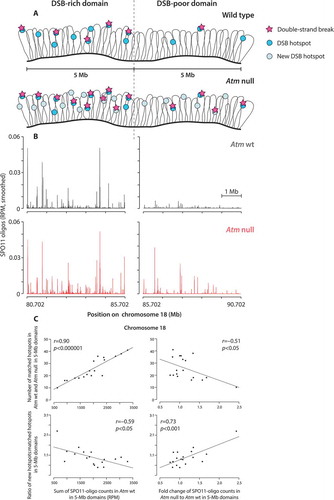
Figure 4. Hypothetical model for ATM-mediated DSB control. (top) Chromatin loops containing DSB hotspots are anchored to the chromosome axis. Before DSB formation, the IHO1-REC114-MEI4 complex and the axis-structural component HORMAD1 assemble along the chromosome axis. The IHO1-REC114-MEI4 complex together with HORMAD1 promotes DSB formation by SPO11. At present, physical interactions between IHO1, REC114, and MEI4 are inferred only from yeast two-hybrid analyzes [Citation118,Citation120], but further support for these interactions comes from extensive two-hybrid and coimmunoprecipitation studies of the orthologous proteins in budding and fission yeasts: Mer2-Rec114-Mei4 [Citation142–Citation145] and Rec15-Rec7-Rec24 [Citation146,Citation147], respectively. (bottom) A DSB is formed preferentially at a DSB hotspot in a chromatin loop tethered to the axis. In response to the DSB, ATM is activated and phosphorylates REC114, HORMAD1, and/or other proteins. Phosphorylation events separately or in combination inhibit additional DSB formation at the same chromatin loop (1) or at the adjacent loop (2) which is pre-activated for DSB formation, by inhibiting or destabilizing of the IHO1-REC114-MEI4 complex. HORMAD1 phosphorylation in the vicinity of the DSB prevents the assembly of the IHO1-REC114-MEI4 complexes and thus DSB formation at other nearby chromatin loop (3)
![Figure 4. Hypothetical model for ATM-mediated DSB control. (top) Chromatin loops containing DSB hotspots are anchored to the chromosome axis. Before DSB formation, the IHO1-REC114-MEI4 complex and the axis-structural component HORMAD1 assemble along the chromosome axis. The IHO1-REC114-MEI4 complex together with HORMAD1 promotes DSB formation by SPO11. At present, physical interactions between IHO1, REC114, and MEI4 are inferred only from yeast two-hybrid analyzes [Citation118,Citation120], but further support for these interactions comes from extensive two-hybrid and coimmunoprecipitation studies of the orthologous proteins in budding and fission yeasts: Mer2-Rec114-Mei4 [Citation142–Citation145] and Rec15-Rec7-Rec24 [Citation146,Citation147], respectively. (bottom) A DSB is formed preferentially at a DSB hotspot in a chromatin loop tethered to the axis. In response to the DSB, ATM is activated and phosphorylates REC114, HORMAD1, and/or other proteins. Phosphorylation events separately or in combination inhibit additional DSB formation at the same chromatin loop (1) or at the adjacent loop (2) which is pre-activated for DSB formation, by inhibiting or destabilizing of the IHO1-REC114-MEI4 complex. HORMAD1 phosphorylation in the vicinity of the DSB prevents the assembly of the IHO1-REC114-MEI4 complexes and thus DSB formation at other nearby chromatin loop (3)](/cms/asset/16f835dd-3a6b-47f4-a37b-4c49d6e88f94/kccy_a_1464847_f0004_oc.jpg)
