Figures & data
Figure 1. Replication-dependent chromatin assembly promotes efficient activation of Chk2 but has minimal effect on γH2AX. (a) HeLa cells transduced with lentiviruses expressing scrambed shRNA or shRNAs against both ASF1A and ASF1B for 72 hours were treated with 40 μg/ml bleomycin for 2 hours, following by washing out the bleomycin at time 0. Samples were taken at the indicated time points after washing out the bleomycin and analyzed by western blotting with the indicated antibodies. Numbers indicate the normalized changes in γH2AX, normalized to GAPDH, and to 1 at time = 0 for each set of lanes within the brackets. (b) As in A, but with analysis by immunofluorescence of γH2AX. (c) As in A, with analysis of Chk2 phosphorylation. Numbers indicate the normalized changes in Chk2p, normalized to Chk2, and to 1 at time = 4. (d) As in B, but with knockdown of CAF-1 p150. (e) As in C, but with knockdown of HIRA. Numbers indicate the normalized changes, as described in D. (f). As in A, but with ASF1A/B and CAF-1 depletion and analysis of KAP-1 phosphorylation.
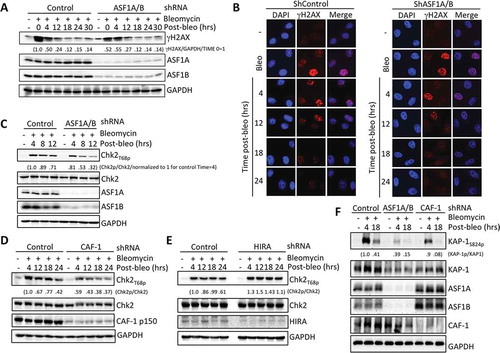
Figure 2. Replication-dependent chromatin assembly promotes efficient activation of ATM. (a) ASF1A/B promotes DSB induced ATMS1981p foci formation. Cells were treated as in , followed by staining for ATMS1981p. Squares indicate areas shown enlarged in the insets. (b) Quantitation of (a). Over 100 cells per sample per experiment were randomly picked to determine the number of foci. Average and SD for 3 independent experiments are shown. (c) Experiment was performed as in , followed by western blotting with the indicated antibodies. (d) As in C, but for HIRA depletion.
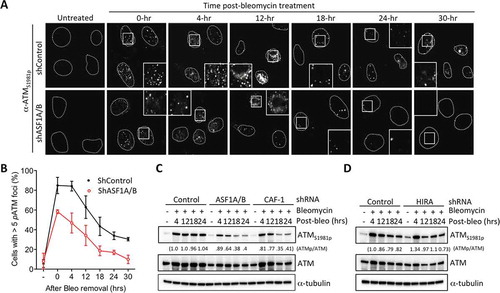
Figure 3. Replication-dependent chromatin assembly promotes H4 K16 acetylation and ASF1A binds hMOF. (a) ASF1A/B were depleted from HeLa cells with shRNAs or siRNAs, followed by western blotting with the indicated antibodies. (b) shRNA depletion of ASF1A or ASF1B alone, as indicated. In the case of ASF1A depletion, either an empty vector or a plasmid expressing H1-ASF1A was reintroduced. (c) hMOF interacts with ASF1A. Immunoprecipitation of hMOF via a FLAG epitope. * refers to a non-specific band running slightly faster than FLAG-hMOF, most likely an antibody chain. Some of the HA-ASF1A in the IP appears to have undergone degradation to a smaller band. (d) Experiment was performed as in with the indicated shRNA depletions followed by western blotting with the indicated antibodies. (e) As in D upon HIRA depletion.
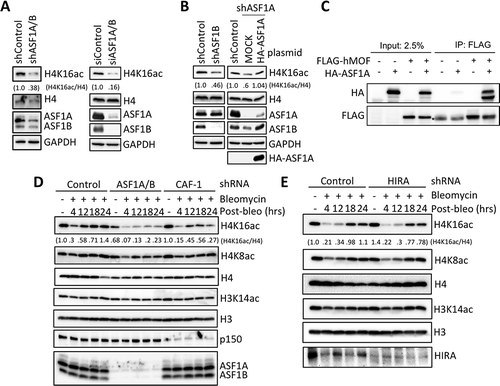
Figure 4. Enhanced DNA-PKcs activation in response to DSBs upon ASF1A/B depletion. (a) Immunofluorescence staining of active DNA-PKcs at the indicated time after washing out 50 μg/ml Bleomcyin upon ASF1A/B depletion by shRNA. (b) The experiment was performed as in , with the indicated shRNA depletions followed by western blotting with the indicated antibodies. (c) The experiment was performed as in , with the indicated shRNA depletions, and fractionation into cytoplasmic, nuclear and chromatin fragments, followed by western blotting with the indicated antibodies.
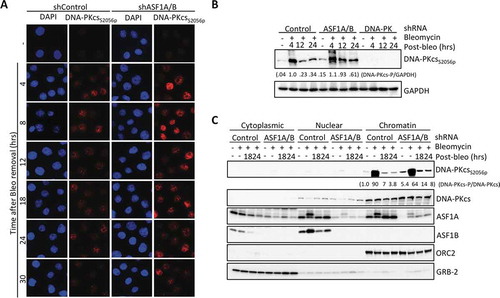
Figure 5. Depletion of ASF1A leads to enhanced levels of DNA-PKcs to DSBs during NHEJ. (a) Cutting and NHEJ repair of the inducible I-PpoI site within the SLCO5A gene is unaffected by ASF1A depletion. Real-time PCR analysis over the break was used to measure the intactness of the I-PpoI site and values were normalized to GAPDH in each sample. Average and SEM are plotted for three individual experiments. (b) ChIP analysis of DNA-PKcsS2056p adjacent to the I-PpoI lesion determined from the same time course shown in A. Representative results are shown.
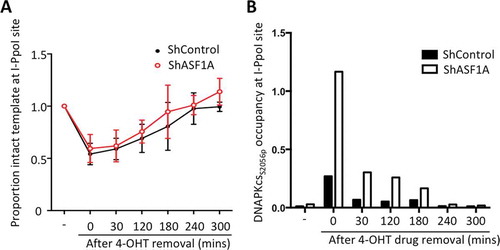
Table 1. A list of antibodies used in this study.
Table 2. A list of siRNAs, shRNAs and gRNA used in this study.
