Figures & data
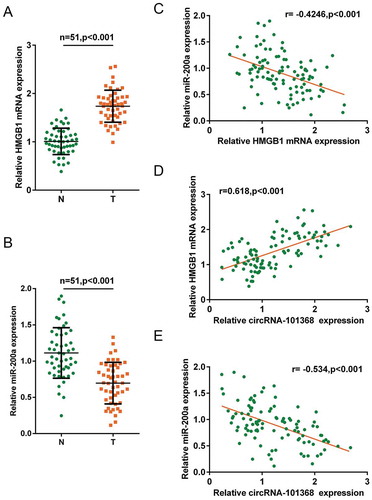
Figure 1. Selection of HCC-associated circRNA and miRNA (a) A total of 366 dysregulated circRNAs in HCC cell lines obtained a fold-change of > 1.5 were shown (GSE78520); the red line indicated that 13 circRNAs obtained a fold-change > 3.0. (b) A total of 142 miRNAs (227 data with significant difference) were differentially expressed in metastatic HCC cell lines compared to those in primary HCC cell lines (GSE43445). (c) Of 205 miRNAs predicted by miRWalk to target HMGB1, 9 were in the above 142 differentially expressed miRNAs (miR-200a, let-7g, miR-193b, let-7b, let-7a, miR-224, miR-532, miR-186 and miR-34a). (d) The expression of circRNA 101368 in tumor and non-cancerous tissue specimens was examined.
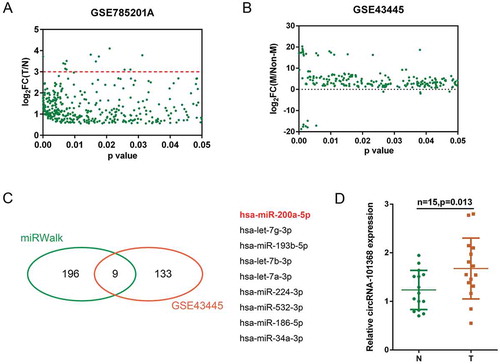
Table 1. Correlation of the expression of circRNA101368 with clinicopathologic features.
Table 2. Univariate and multivariate analysis for factors related to overall survival using the COX proportional hazard model.
Figure 2. circRNA 101368 is overexpressed in HCC tissues and cell lines and is correlated with poorer prognosis (a) Expression of circRNA 101368 in 51 paired tumor and non-cancerous tissue specimens was examined using real-time PCR. (b) Expression of circRNA 101368 in 51 cases of HCC tissues was analyzed according to TNM stages. (c) The fold-change of circRNA 101368 expression in 51 cases of HCC tissues was shown as log2 (tumor/normal). (d) Kaplan-Meier overall survival curves for 51 cases grouped by circRNA 101368 expression. (e) circRNA 101368 expression in five HCC cell lines and a normal hepatocyte was examined.
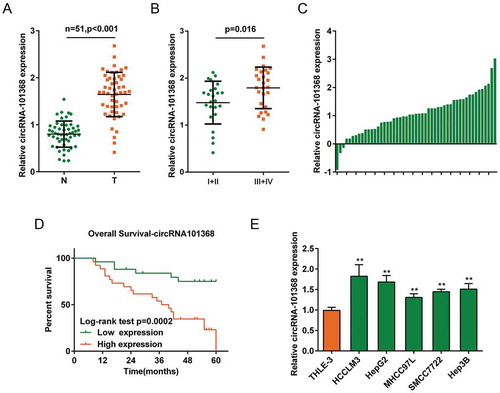
Figure 3. circRNA 101368 knockdown suppressed the cell migration and HMGB1/RAGE signaling in HCC cell lines (a) circRNA 101368 knockdown in HepG2 and HCCLM3 cell lines was achieved by transfection of si-circRNA 101368–1 or si-circRNA 101368–2, as confirmed using real-time PCR. (b-c) The cell migration of HepG2 and HCCLM3 cell lines transfected with si-circRNA 101368–1 or si-circRNA 101368–2 was examined using Transwell assays. (d-e) The protein levels of HMGB1, RAGE, NF-κB and E-Cadherin in HepG2 and HCCLM3 cell lines transfected with si-circRNA 101368 were examined using Immunoblotting assays.
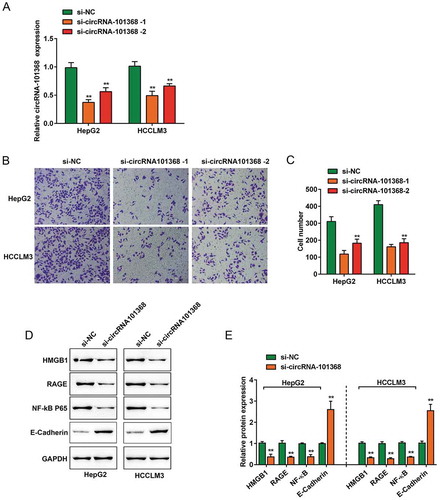
Figure 4. miR-200a is a direct downstream target of circRNA 101368 (a) HepG2 and HCCLM3 cell lines were transfected with miR-200a mimics or miR-200a inhibitor to achieve miR-200a expression, as confirmed using real-time PCR; (b) transfected HCC cell lines were examined for circRNA 101368 expression. (c) HepG2 and HCCLM3 cell lines were transfected with si-circRNA 101368 and examined for miR-200a expression. (d) The miR-200a binding site in circRNA 101368 predicted by lncTar was shown. A wild-type and a mutant-type circRNA 101368 luciferase reporter vector were constructed, containing wild-type or mutant-type miR-200a binding site, respectively. (e) HEK293 cells were co-transfected with the above luciferase reporter vectors and miR-200a mimics/inhibitor and examined for luciferase activity. (f-g) Association of circRNA 101368, miR-200a and HMGB1 with AGO2. Detection of AGO2 and IgG using Immunoblotting and detection of circRNA 101368, miR-200a and HMGB1 using real-time PCR.
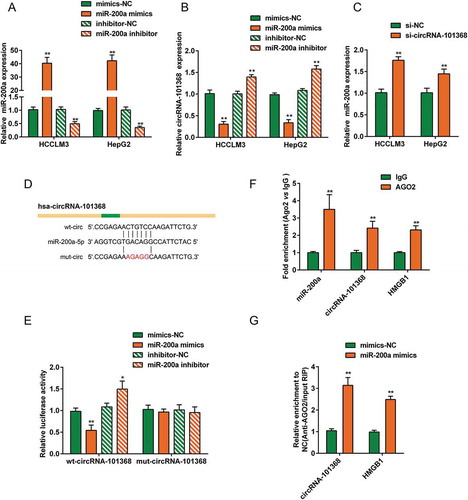
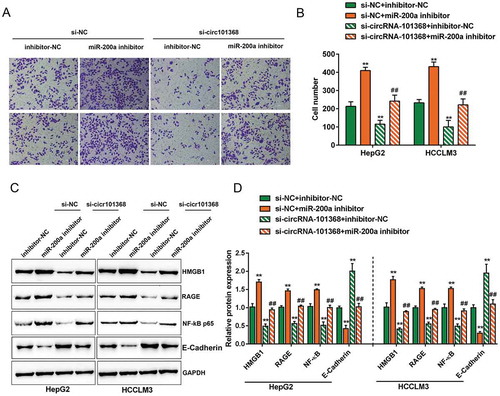
Figure 5. The effect of circRNA 101368 on HCC cell migration could be partially reversed by miR-200a through HMGB1/RAGE signaling HepG2 and HCCLM3 cell lines were co-transfected with si-circRNA 101368 and miR-200a inhibitor and examined for cell migration (a-b) and protein levels of HMGB1, RAGE, NF-κB (p65) and E-Cadherin (c-d) .