Figures & data
Figure 1. Down-regulated miR-16 expression in OSCC tissues correlated with OSCC aggressiveness. (a) The relative expression of miR-16 in OSCC tissues (n = 131) and adjacent normal tissues (n = 131) as determined by qRT-PCR. Tumor: OSCC tissues; Non-tumor: adjacent normal tissues. (b) The expression levels of miR-16 in five OSCC cell lines and HOK were showed. HOK: human oral keratinocytes; (c) Comparison of miR-16 expressions between non-metastatic (n = 76) and metastatic (n = 55) OSCC tissues. (d) Kaplan–Meier analysis of the overall survival of OSCC patients with different miR-16 expression in tumor tissues. (e) Kaplan-Meier analysis for overall survival of 521 OSCC patients by log-rank test according to different miR-16 levels based on TCGA database. *p < 0.05, **p < 0.01, ***p < 0.001 vs. Control group.
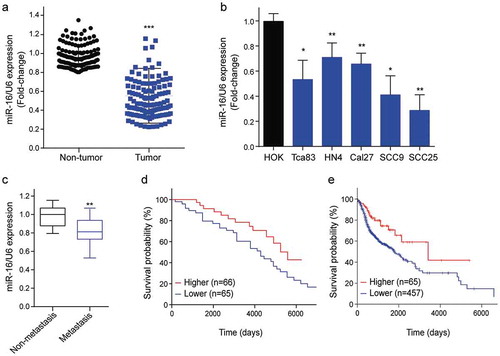
Table 1. Correlation between clinical pathologic features and the miR-16 expressions in patients with oral squamous cell carcinoma.
Table 2. Univariable and multivariable analyses of factors and overall survival of patients with OSCC.
Table 3. Correlations between the levels of miR-16 and the mRNA expression of TLK1 in 131 patients with OSCC.
Figure 2. TLK1 was upregulated in OSCC tissues and cells. (a) Representative images of TLK1 expression in OSCC tissues and adjacent normal tissues by immunohistochemistry (IHC) staining. (b) Relative TLK1 mRNA expressions in five OSCC cell lines and HOK cells. (c) The protein levels of TLK1 in five OSCC cell lines were analyzed by Western blot. (d) Kaplan-Meier analysis for overall survival of 495 OSCC patients by log-rank test according to different TLK1 levels based on TCGA database. **p < 0.01, ***p < 0.001 vs. HOK.
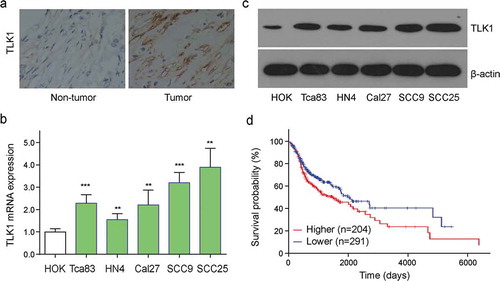
Figure 3. TLK1 is a potential target of miR-16 in OSCC cells. (a) miR-16 was identified to potentially interact with the 3-UTR of TLK1 (Wild type). The putative miR-16 binding sites were mutated to generate mutated TLK1 3-UTR (Mutant). (b) Luciferase reporter assay was performed to determine the interaction between miR-16 and the 3`-UTR of TLK1 mRNA in SCC9 and SCC25 cells. SCC9 cells were transfected with lentivirus vector-expressing miR-16 (miR-16) or its negative control (Vector). (c) Analysis of TLK1 mRNA expression by qRT-PCR. (d) After transfection, SCC9 and SCC25cells were subjected to western blot for the level of TLK1 protein. (e) Immunofluorescence staining of TLK1 in SCC9 cells with miR-16 overexpression. **p < 0.01, ***p < 0.001 vs. Vector. The experiments were performed in triplicate.
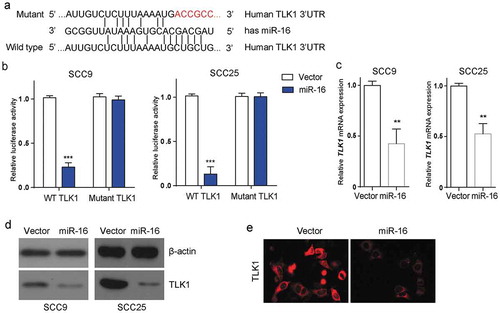
Figure 4. Forced expression of miR-16 suppresses OSCC progression in vitro and in vivo. SCC9 and SCC25 cells were transfected with miR-16-contained lentivirus vector (miR-16) or its negative control (Vector). (a) The expression levels of miR-16 were detected after transfection in SCC9 and SCC25 cells. (b) Colony formation ability and (c) cell viability were examined in miR-16- and vector- SCC9 and SCC25 cells. (d, e) BALB/c nude mice were randomly divided into 2 groups with 6 mice in each group. MiR-16-overexpressed SCC9 cells (miR-16) or vector cells (Vector) were injected into the nude mice, respectively. (d) Tumor volume was measured every 3 days starting at the third day after tumor cell injection. (e) Representative images and quantification of H&E staining of metastatic nodes in lung tissue. *p < 0.05, **p < 0.01, vs. Vector.
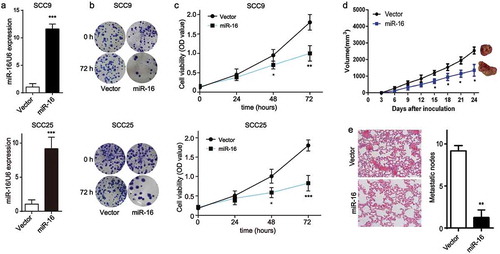
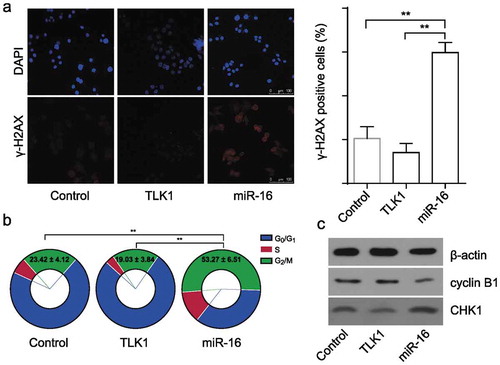
Figure 5. Overexpression of TLK1 reverses miR-16-mediated DNA damage and G2/M arrest. (a) Representative images of γ-H2AX foci in miR-16-overexpressed (miR-16), TLK1-overexpressed or vector SCC9 cells by immunofluorescence staining. Quantification of the number of γ-H2AX foci in miR-16-overexpressed (miR-16), TLK1-overexpressed or vector SCC9 cells. (b) Flow cytometry analysis of cell cycle in miR-16-overexpressed (miR-16), TLK1-overexpressed or vector SCC9 cells. (c) Western blot analysis of cell cycle associated proteins in miR-16-overexpressed (miR-16), TLK1-overexpressed or vector SCC9 cells. **p < 0.01 vs. Vector. The experiments were performed in triplicate.